
Next: Acknowledgment
Up: Steered Molecular Dynamics
Previous: Constant Force Pulling
Subsections
Analysis of Results
Hopefully your constant velocity SMD simulation has finished successfully and you can proceed with analyzing the generated data, specifically the trajectory and the force applied to the SMD atom. In case you have not succeeded with your simulations we provide the respective files in the subdirectories example-output. For instance, if you do not find a file in the directory common, you should find it in the directory common/example-output.
- 1
- In VMD choose the New Molecule... menu item. Using the Browse... and the Load buttons, load the file ubq.psf in the common directory.
- 2
- In the Molecule File Browser window, use the Browse... and the Load buttons load the trajectory file 3-1-pullcv/ubq_ww_pcv.dcd. Close the Molecule File Browser.
In the screen you should be able to see the trajectory of the molecular dynamics simulation.
By choosing an appropriate representation (e.g. New Cartoon) look how the 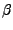 strands behave along the trajectory and how one end of the protein remains fixed while the other one is pulled as you had set it up (Fig. 21). Note that secondary structure will be calculated for the New Cartoon representation based on the frame of the trajectory you are viewing. If you move to another trajectory frame and would like to view the protein's secondary structure in that frame, type mol ssrecalc top in the TkCon window and VMD will update it.
strands behave along the trajectory and how one end of the protein remains fixed while the other one is pulled as you had set it up (Fig. 21). Note that secondary structure will be calculated for the New Cartoon representation based on the frame of the trajectory you are viewing. If you move to another trajectory frame and would like to view the protein's secondary structure in that frame, type mol ssrecalc top in the TkCon window and VMD will update it.
Figure:
Snapshots of ubiquitin pulling with constant velocity at three different time steps.
![\begin{figure}\begin{center}
\par
\par
\latex{
\includegraphics[scale=0.5]{pictures/tut_unit03_pcvt}
}
\end{center}
\end{figure}](img199.png) |
After inspecting your trajectory you should extract the force applied to the SMD atom from the NAMD output file:
- 3
- In the VMD TkCon window, set your current directory to 3-1-pullcv by typing: cd 3-1-pullcv
- 4
- In directory 3-1-pullcv, create the subdirectory called analysis by typing: mkdir analysis
You will now use a script designed to remove the necessary information from the NAMD log file and calculate the force of pulling. The NAMD log file provides SMD information on lines beginning with the tag ``SMD": the current timestep, current position of the center of mass of the restrained atoms, and the current force applied to the center of mass (in piconewtons).
- 5
- Source the provided tcl script for getting force information from the NAMD log file. Type the following command in the VMD TkCon window:
In order to obtain the force in the direction of pulling you need to calculate
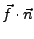 , where
, where 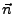 is the normalized direction of pulling (in our example it was 0.443, 0.398, 0.803). The script will ask you for these values. Simply enter them as n_x, n_y, and n_z. The script will create a file ft.dat (in the analysis directory) which contains just the simulation timestep and the magnitude of the force on the dummy atom at that time.
is the normalized direction of pulling (in our example it was 0.443, 0.398, 0.803). The script will ask you for these values. Simply enter them as n_x, n_y, and n_z. The script will create a file ft.dat (in the analysis directory) which contains just the simulation timestep and the magnitude of the force on the dummy atom at that time.
- 6
- The contents of the script are shown below. You should understand what each line does in order to make tcl scripting a part of your repertoire.
### Open the log file for reading and the output .dat file for writing
set file [open ubq_ww_pcv.log r]
set output [open analysis/ft.dat w]
### Gather input from user.
puts "Enter a value for n_x:"
set nx [gets stdin]
puts "Enter a value for n_y:"
set ny [gets stdin]
puts "Enter a value for n_z:"
set nz [gets stdin]
### Loop over all lines of the log file
set file [open ubq_ww_pcv.log r]
while { [gets $file line] != -1 } {
### Determine if a line contains SMD output. If so, write the
### timestep followed by f(dot)n to the output file
if {[lindex $line 0] == "SMD"} {
puts $output "[lindex $line 1] [expr $nx*[lindex $line 5]
+ $ny*[lindex $line 6] + $nz*[lindex $line 7]]"
}
}
### Close the log file and the output .dat file
close $file
close $output
- 7
- Type xmgrace analysis/ft.dat. A plot of force versus time should appear on your screen (Fig. 22).
- 8
- In VMD, look again at the trajectory at different times and try to identify the events that are related to peaks in the force. When you are done close xmgrace.
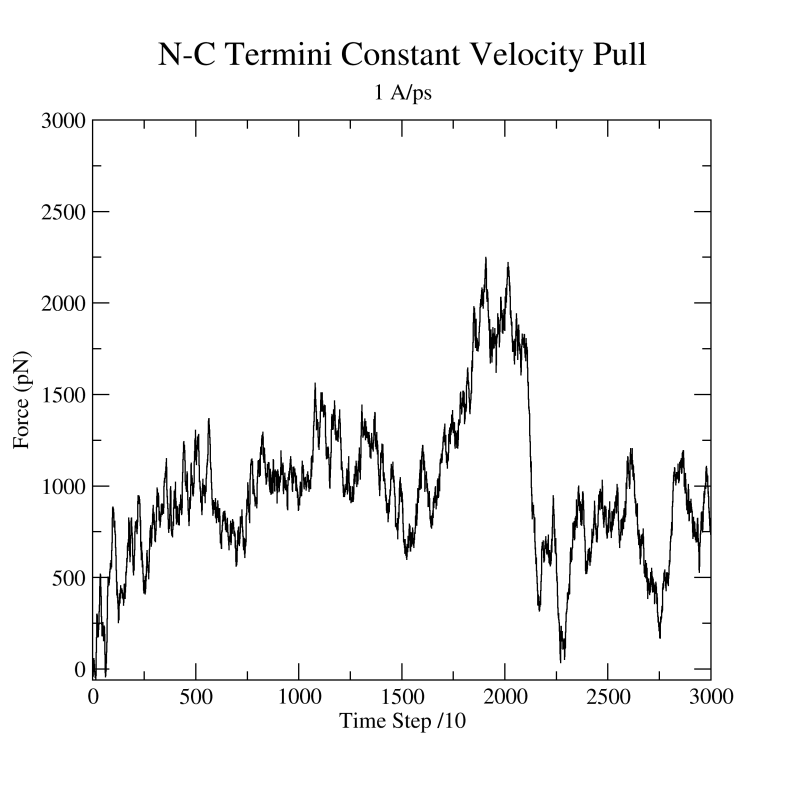
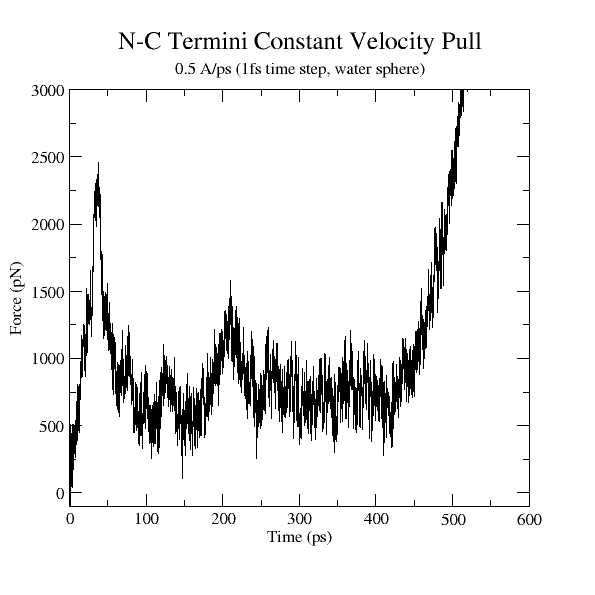
Figure 22:
Force vs Time. Your plot should resemble the graph on the left. The graph on the right was obtained in a simulation where ubiquitin was placed in a water sphere (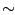 26000 atoms) and pulled at 0.5 Å/ps using a
time step of 1 femtosecond. The results of this last simulation are in better agreement with actual AFM experiments. The force
at the end (
26000 atoms) and pulled at 0.5 Å/ps using a
time step of 1 femtosecond. The results of this last simulation are in better agreement with actual AFM experiments. The force
at the end (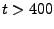 ps) increases when the protein becomes completely unfolded. The first peak in the force is associated with the breaking of hydrogen bonds between two
ps) increases when the protein becomes completely unfolded. The first peak in the force is associated with the breaking of hydrogen bonds between two 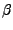 strands.
strands.
|
|
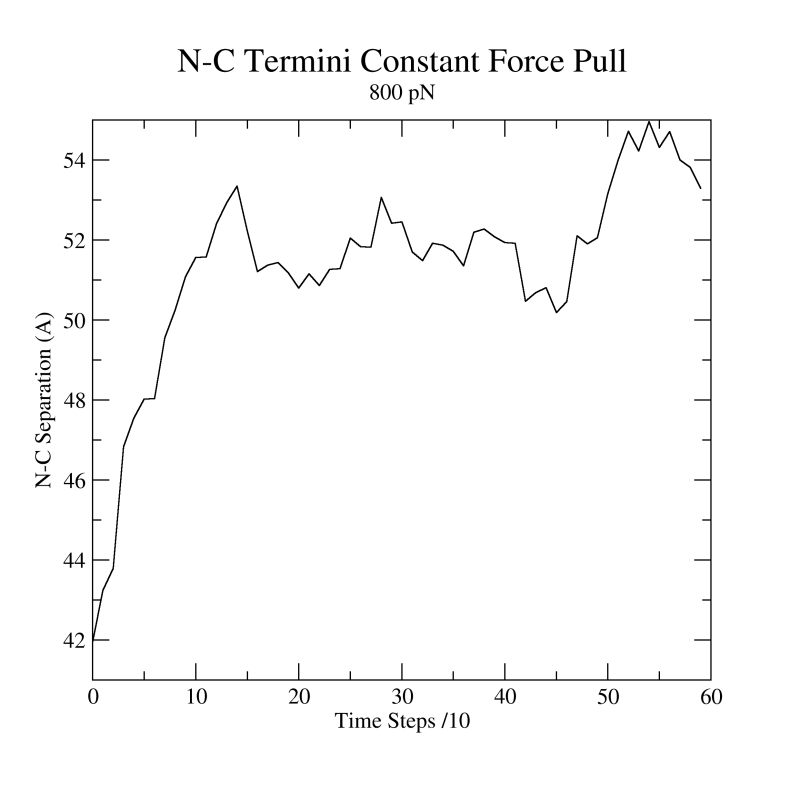
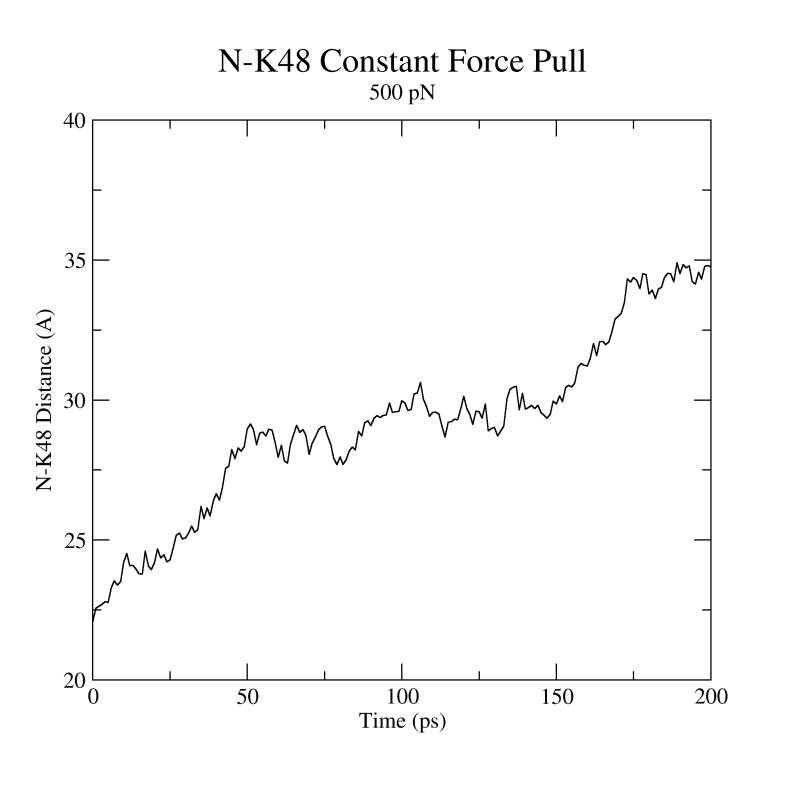
Figure 23:
Distance vs Time Steps. Your plot should resemble the graph on the left. The graph on the right was obtained in a simulation where ubiquitin was placed in a water box and pulled from the C-Terminus while keeping the residue K48 fixed. In this case we can observe an intermediate state.
|
|
- 1
- In VMD, delete the current molecule by choosing the Molecule
 Delete Molecule menu item.
Delete Molecule menu item.
- 2
- Choose the New Molecule... menu item. Using the Browse... and the Load buttons load the file ubq.psf located in the common directory.
- 3
- In the Molecule File Browser window, use the Browse... and the Load buttons load the trajectory file 3-2-pullcf/ubq_ww_pcf.dcd. As mentioned before, in case that you have not accomplished the simulation to generate the file, we provide such file at 3-2-pullcf/example-output/ubq_ww_pcf.dcd.
In the OpenGL Display window you will see the trajectory of the constant force pulling simulation.
- 4
- Once the complete trajectory is loaded, use the slider in the VMD Main window to go back to the first frame.
- 5
- Choose the Graphics
 Representations... menu item.
In the Graphical Representations window choose the New Cartoon drawing method.
Representations... menu item.
In the Graphical Representations window choose the New Cartoon drawing method.
- 6
- Press the Create Rep button in order to create a new representation. In the
Selected Atoms text entry delete the word all and type protein and resid 1 76 and name CA.
- 7
- For this representation choose the VDW drawing method. You should be able to see
the two C
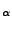 atoms at the end of your protein as spheres.
atoms at the end of your protein as spheres.
- 8
- Hide the first representation by double clicking on it in the Graphical Representations window.
- 9
- Click in the OpenGL Display window and press the key 2 (Shortcut to bond labels). Your
cursor should look like a cross. Pick the two spheres displayed. A line linking both atoms should appear with the distance between them displayed.
- 10
- Choose the Graphics
 Labels... menu item. In the Labels window, choose the label type Bonds. Select the bond displayed, click on the Graph tab and then click on the Graph button. This will create a plot of the distance between these two atoms over time (Fig. 23). You also have the option to write the data and to a file, which can then be read by an external graphing program.
Labels... menu item. In the Labels window, choose the label type Bonds. Select the bond displayed, click on the Graph tab and then click on the Graph button. This will create a plot of the distance between these two atoms over time (Fig. 23). You also have the option to write the data and to a file, which can then be read by an external graphing program.
This is the end of the NAMD tutorial. We hope you can now make use of VMD and NAMD in your work.


Next: Acknowledgment
Up: Steered Molecular Dynamics
Previous: Constant Force Pulling
namd@ks.uiuc.edu
![]() strands behave along the trajectory and how one end of the protein remains fixed while the other one is pulled as you had set it up (Fig. 21). Note that secondary structure will be calculated for the New Cartoon representation based on the frame of the trajectory you are viewing. If you move to another trajectory frame and would like to view the protein's secondary structure in that frame, type mol ssrecalc top in the TkCon window and VMD will update it.
strands behave along the trajectory and how one end of the protein remains fixed while the other one is pulled as you had set it up (Fig. 21). Note that secondary structure will be calculated for the New Cartoon representation based on the frame of the trajectory you are viewing. If you move to another trajectory frame and would like to view the protein's secondary structure in that frame, type mol ssrecalc top in the TkCon window and VMD will update it.
![]() , where
, where ![]() is the normalized direction of pulling (in our example it was 0.443, 0.398, 0.803). The script will ask you for these values. Simply enter them as n_x, n_y, and n_z. The script will create a file ft.dat (in the analysis directory) which contains just the simulation timestep and the magnitude of the force on the dummy atom at that time.
is the normalized direction of pulling (in our example it was 0.443, 0.398, 0.803). The script will ask you for these values. Simply enter them as n_x, n_y, and n_z. The script will create a file ft.dat (in the analysis directory) which contains just the simulation timestep and the magnitude of the force on the dummy atom at that time.