Nanosensors Spotlights

image size:
1.7MB
made with VMD
DNA sequencing is achieved by following a strand of DNA at a speed that permits recognition of the DNA bases in their actual order, thousands of bases or more for each pass. Nanotechnology can assist in the task, in principle, by furnishing sensors that can resolve single DNA bases and nano-mechanical actuators that pull the DNA in a controlled fashion passing through the sensor. Instead of building and testing actual sensors and actuators it is cheaper and faster to simulate them. Nanoengineers have indeed succeeded with such simulations over the last decade focussing on silicon technology (see October 2004 highlight: Transistor Meets DNA). Now the engineers have moved with their simulations to graphene technology that promises much better resolving power as sensors are thinner and as signals can be detected electronically in graphene (December 2013 and November 2014 highlights). The main unsolved problem is the mechanical actuator: how can one control movement of DNA through a graphene sensor such that measured signals become less noisy and bases can be recognized? A recent study, based on molecular dynamics simulations with NAMD and quantum electronics calculations of graphene, suggests use of an actuator that simultaneously stretches the DNA and pulls it through the sensor. This manipulation leads to a stepwise translocation of DNA through the graphene nanosensor, slowing down DNA translocation and stabilizing DNA bases inside the sensor. Read more on our graphene nanopore website.
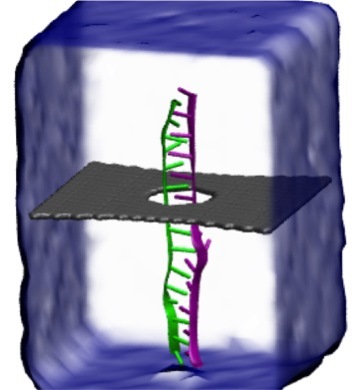
image size:
287.1KB
made with VMD
Threading DNA through a nanometer-size pore, so called nanopores, drilled into an ultrathin graphene membrane is a promising approach to build nanobiosensors for sequencing the human genome. Graphene nanopores can detect translocating DNA by recording concomitant flow of charged ions through the pore (see December 2011 highlight). As reported in the December 2013 highlight, graphene, which is an electrical conductor, offers a new way of sensing DNA molecules by monitoring sheet currents along the graphene membrane. DNA is a highly extensible molecule and upon mechanical manipulation can change its structure from a canonical helical conformation to a linear zipper-like conformation. A new study, which combines classical molecular dynamics simulations using NAMD with quantum mechanical simulations, suggests that sheet currents, in graphene membranes, can be used to detect conformation and sequence of a DNA molecule passing through the nanopore. This new research will guide the development of graphene-based nanosensors for DNA detection. More information can be found on our graphene nanopore website.
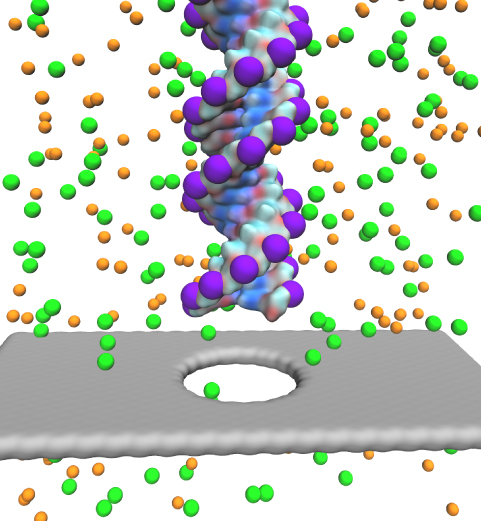
image size:
210.7KB
made with VMD
Characterizing the genetic makeup of individuals can help select the best treatment for individuals, but requires that sequencing of the whole DNA of patients can be achieved for less than $1000. Fortunately, recent discoveries in physics promise help. Indeed, the discovery of the thinnest material known to mankind, graphene, promises a new and cheaper way to sequence human DNA. As reported in the December 2011 highlight, graphene pores can conduct electrophoretically DNA through very small pores, so-called nanopores. A new study has demonstrated that graphene, forming a single atomic layer thin stripe with a nanopore in the middle, can conduct an electrical current, the sheet current, around the pore. The sheet current is sensitive to the DNA passing through the pore and may even sense the passing DNA's sequence. In this case monitoring the current can establish a DNA sequence reader. The sensitivity of the sheet current depends critically on an orderly passage of DNA. Optimal sensitivity can arise when the passing DNA is stretched mechanically. Molecular dynamics simulations using NAMD suggest conditions for mechanically manipulating DNA for optimal sequence analysis. More on our graphene nanopore website.
Threading DNA electrically through nanometer-sized pores, so-called nanopores, holds promise for detecting and sequencing DNA (see Nov 2005 and Oct 2004 highlights). Nanopore measurements tend to be the more sensitive the smaller the pores are. The material graphene, which is just one atom thick and looks like a two-dimensional ``honeycomb" made up of carbon atoms, offers the ultimate physical resolution for measuring DNA (the stacking distance between base-pairs in DNA is about 0.35 nm). As reported recently, molecular dynamics simulations using NAMD revealed the motion of DNA being threaded through graphene nanopores at atomic level resolution. Simulations not only agree qualitatively with previous experiments on DNA translocation through graphene nanopores, but go one step further than the experiments and suggest how individual base pairs can be discriminated. The recent computational study is one further example for the guidance that molecular dynamics simulations provide in nanosensor development (see a recent review). More information can be found on our graphene nanopore website.




