Viruses
Viruses are small intracellular parasites that invade the cells of virtually all known organisms. They reproduce by utilizing the cell's machinery to replicate viral proteins and genomic material, generally damaging or killing the host cell in the process; subsequentelly, a large number of newly generated viruses go on to infect other cells. Viruses are responsible for a wide variety of human diseases, ranging from the common (influenza and colds) to the exotic (AIDS, West Nile virus and Zika). Some viruses which are not dangerous to humans can also be exploited in technological applications, in addition, viruses find use in genetic engineering applications and increasingly in the design of new nanomaterials. At the very least, all viruses contain two components: the capsid (a protein shell), and a genome, consisting of either DNA or RNA. Some viruses also include accessory proteins to aid in infection, and in some cases a lipid bilayer to further protect their contents from the environment. The viral life cycle itself is deceivingly simple: viruses enter the cell, typically (but not always) through the interaction of their capsid with a receptor on the cell surface; the virus must then somehow disassemble its capsid to release its genetic material and any necessary helper proteins. The viral genome is then replicated and the proteins it codes for are synthesized to produce the raw material for the production of new viral particles; these new viruses then assemble and bud from the cell either through the membrane or upon cell death.
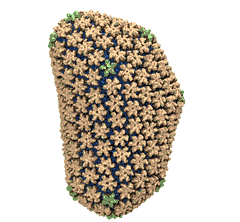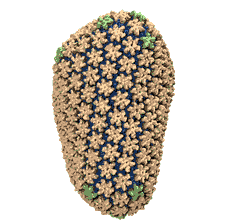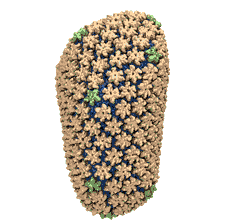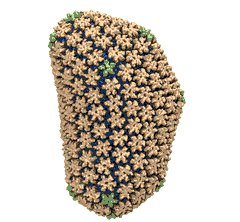
Spotlight: Elusive HIV-1 Capsid (Jun 2013)
Human immunodeficiency virus type 1 (HIV-1) is the major cause of AIDS, for which treatments need to be developed continuously as the virus becomes quickly resistant to new drugs. When the virus infects a human cell it releases into the cell its capsid, a closed, stable container protecting the viral genetic material. However, interaction with the cell triggers at some point an instability of the capsid, leading to a well timed release of the genetic material that merges then with the cell's genes and begins to control the cell. The dual role of the capsid, to be functionally both stable and unstable, makes it in principle an ideal target for antiviral drugs and, in fact, treatments of other viral infections successfully target the respective capsids. The size of the HIV-1 capsid (about 1,300 proteins), and its irregular shape had prevented so far the resolution of a full capsid atomic-level structure. However, in a tour de force effort, groups of experimental and computational scientists have now resolved the capsid's chemical structure (deposited to the protein data bank under the accession codes 3J3Q and 3J3Y). As reported recently (see also journal cover), the researchers combined NMR structure analysis, electron microscopy and data-guided molecular dynamics simulations utilizing VMD to prepare and analyze simulations performed using NAMD on one of the most powerful computers worldwide, Blue Waters, to obtain and characterize the HIV-1 capsid. The discovery can guide now the design of novel drugs for enhanced antiviral therapy. More information is available on our virus website, in video, and in a press release.