TCB Members, NAMD, and VMD Part of 2021 ACM Gordon Bell Finalist for COVID-19 Research
Members of the Theoretical and Computational Biophysics Group were part of a multi-institutional interdisciplinary team awarded as finalists at Supercomputing 2021 with the internationally recognized ACM Gordon Bell Special Prize for COVID-19 Research, for 2021.
Team members from U. Illinois provided key methodological and scalability advances in the NAMD molecular dynamics simulation software, and technological improvements to VMD, a key molecular modeling tool used to prepare, visualize, and analyze FFEA and NAMD simulations of the RTC. The NAMD and VMD software advances provided by U. Illinois team members enabled the science campaign to more efficiently utilize state-of-the-art supercomputers at national computing centers, including NERSC Perlmutter, ALCF ThetaGPU, and TACC Frontera.
These achievements required the unique skills of a diverse multi-institutional team to overcome both technical challenges and an extremely compressed research timeline. Every team member played a vital role in achieving the final outcome.
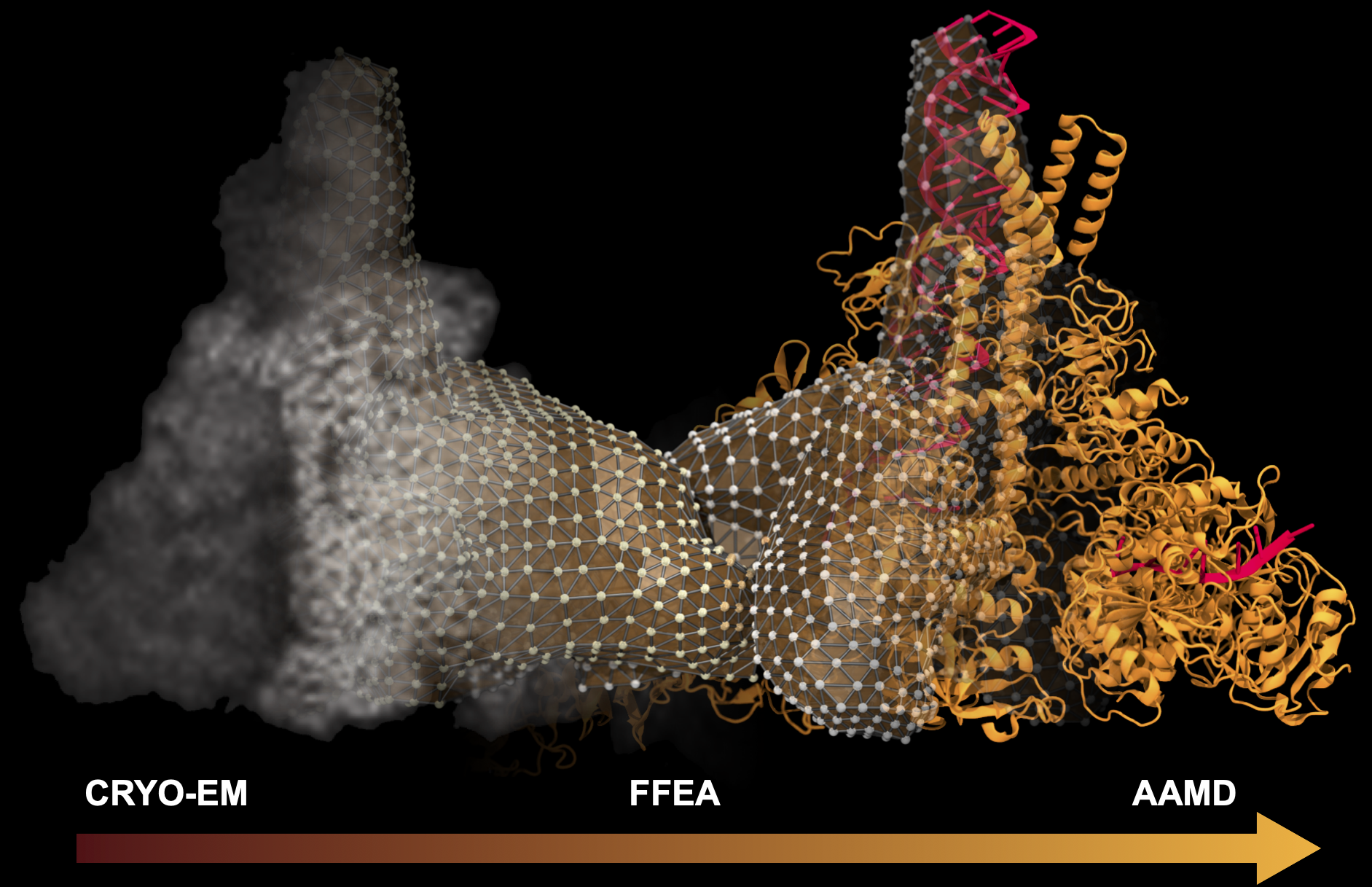
Intelligent Resolution: Integrating Cryo-EM with AI-Driven Multi-Resolution Simulations to Observe the SARS-CoV-2 Replication-Transcription Machinery in Action
Abstract: The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) replication transcription complex (RTC) is a multi-domain protein responsible for replicating and transcribing the viral mRNA inside a human cell. Attacking RTC function with pharmaceutical compounds is a pathway to treating COVID-19. Conventional tools, e.g., cryo-electron microscopy and all-atom molecular dynamics (AAMD), do not provide sufficiently high resolution or timescale to capture important dynamics of this molecular machine. Consequently, we develop an innovative workflow that bridges the gap between these resolutions, using mesoscale fluctuating finite element analysis (FFEA) continuum simulations and a hierarchy of AI-methods that continually learn and infer features for maintaining consistency between AAMD and FFEA simulations. We leverage a multi-site distributed workflow manager to orchestrate AI, FFEA, and AAMD jobs, providing optimal resource utilization across HPC centers. Our study provides unprecedented access to study the SARS-CoV-2 RTC machinery, while providing general capability for AI-enabled multi-resolution simulations at scale.- bioRxiv preprint of finalist research paper, to appear International Journal of High Performance Computing Applications (IJHPCA)
- Supercomputing 2021 team presentation (YouTube)
Team presentation at Supercomputing 2021:


used to refine the cryo-EM structure.

for the 1.1M- and 2.2M-atom RTC. The Black reference
line shows the performance of the 1.1M-atom RTC
on 128 nodes of TACC Frontera

provide insites that are helpful in determining the
intrinsic concerted motions in the RTC that are
implicitly encoded in the experimental data.
- Balsam (GitHub)
- DeepDriveMD (GitHub)
- FFEA (Home Page)
- MDLearn (GitHub)
- NAMD, Charm++ (Home Page)
- VMD (Home Page)
- Nature Computational Science: Fighting COVID-19 with HPC, Dec 13, 2021
- Seimaxim: Researchers Fight COVID, Advance Science with NVIDIA A100 GPU, Nov 23, 2021
- UCL: UCL and UK scientists part of team nominated for supercomputing prize, Nov 18, 2021
- Oak Ridge National Laboratory: Waltzing the virus: Study on COVID-19 reproduction earns Gordon Bell Special Prize nomination, Nov 17, 2021
- OLCF: Waltzing the virus: Study on COVID-19 reproduction earns Argonne researchers Gordon Bell Special Prize nomination, Nov 16, 2021
- Argonne National Laboratory: Waltzing the virus: Study on COVID-19 reproduction earns Argonne researchers Gordon Bell Special Prize nomination, Nov 16, 2021
- Charmworks: Two projects using Charm++ named finalists for Gordon Bell Special Prize, Nov 15, 2021
- HPCWire: Gordon Bell Finalists Fight COVID, Advance Science with NVIDIA Technologies, Nov 15, 2021
- NVIDIA: Gordon Bell Finalists Fight COVID, Advance Science with NVIDIA Technologies, Nov 15, 2021
Research Team:
- Alexander Brace
- Austin Clyde
- Murali Emani
- Ian Foster
- Heng Ma
- Arvind Ramanathan
- Michael Salim
- Rick Stevens
- Anda Trifan
- Venkatram Vishwanath
- Hyenseung Yoo
- Maxim Zvyagin
- David J. Hardy
- Defne Gorgun
- James C. Phillips
- John E. Stone
- Emad Tajkhorshid
- Noah Trebesch
- Anda Trifan
- Zongyi Li
- Sarah A. Harris
- Geoffrey Wells
- Chakra Chennubhotla
- Shantenu Jha
- Jessica Liu
- Tavneer Raza
- Vishal Subbiah
- Anima Anandkumar
- David Clark
- Tom Gibbs
- Venkatesh Mysore
- Aristeidis Tsaris
- Junqi Yin
- Tom Burnley
- Lei Huang
- John McCalpin