Over 5,000 citations of NAMD reference paper
October 15, 2015
The NAMD developers thank our users for 5,000 citations of our 2005 reference paper:
James C. Phillips, Rosemary Braun, Wei Wang, James Gumbart, Emad Tajkhorshid, Elizabeth Villa, Christophe Chipot, Robert D. Skeel, Laxmikant Kale, and Klaus Schulten. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26:1781-1802, 2005.
NAMD is a parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. NAMD scales to hundreds of processors on high-end parallel platforms, as well as tens of processors on low-cost commodity clusters, and also runs on individual desktop and laptop computers. NAMD works with AMBER and CHARMM potential functions, parameters, and file formats. This article, directed to novices as well as experts, first introduces concepts and methods used in the NAMD program, describing the classical molecular dynamics force field, equations of motion, and integration methods along with the efficient electrostatics evaluation algorithms employed and temperature and pressure controls used. Features for steering the simulation across barriers and for calculating both alchemical and conformational free energy differences are presented. The motivations for and a roadmap to the internal design of NAMD, implemented in C++ and based on Charm++ parallel objects, are outlined. The factors affecting the serial and parallel performance of a simulation are discussed. Finally, typical NAMD use is illustrated with representative applications to a small, a medium, and a large biomolecular system, highlighting particular features of NAMD, for example, the Tcl scripting language. The article also provides a list of the key features of NAMD and discusses the benefits of combining NAMD with the molecular graphics/sequence analysis software VMD and the grid computing/collaboratory software BioCoRE. NAMD is distributed free of charge with source code at www.ks.uiuc.edu.
This paper is available from our website or directly from the journal.
The NAMD License Agreement requires that this paper be cited by any published work which utilizes NAMD. Proper citation is essential to continued NIH funding for NAMD development, as it is a primary way in which we demonstrate the value of our software to the scientific community. As of May 2015, over 6000 publications have cited either the 1999 or 2005 reference papers.
Below are a few of the many biomedically, environmentally, and methodologically relevant discoveries made by NAMD users since our September 2014 milestone of 4,000 citations.
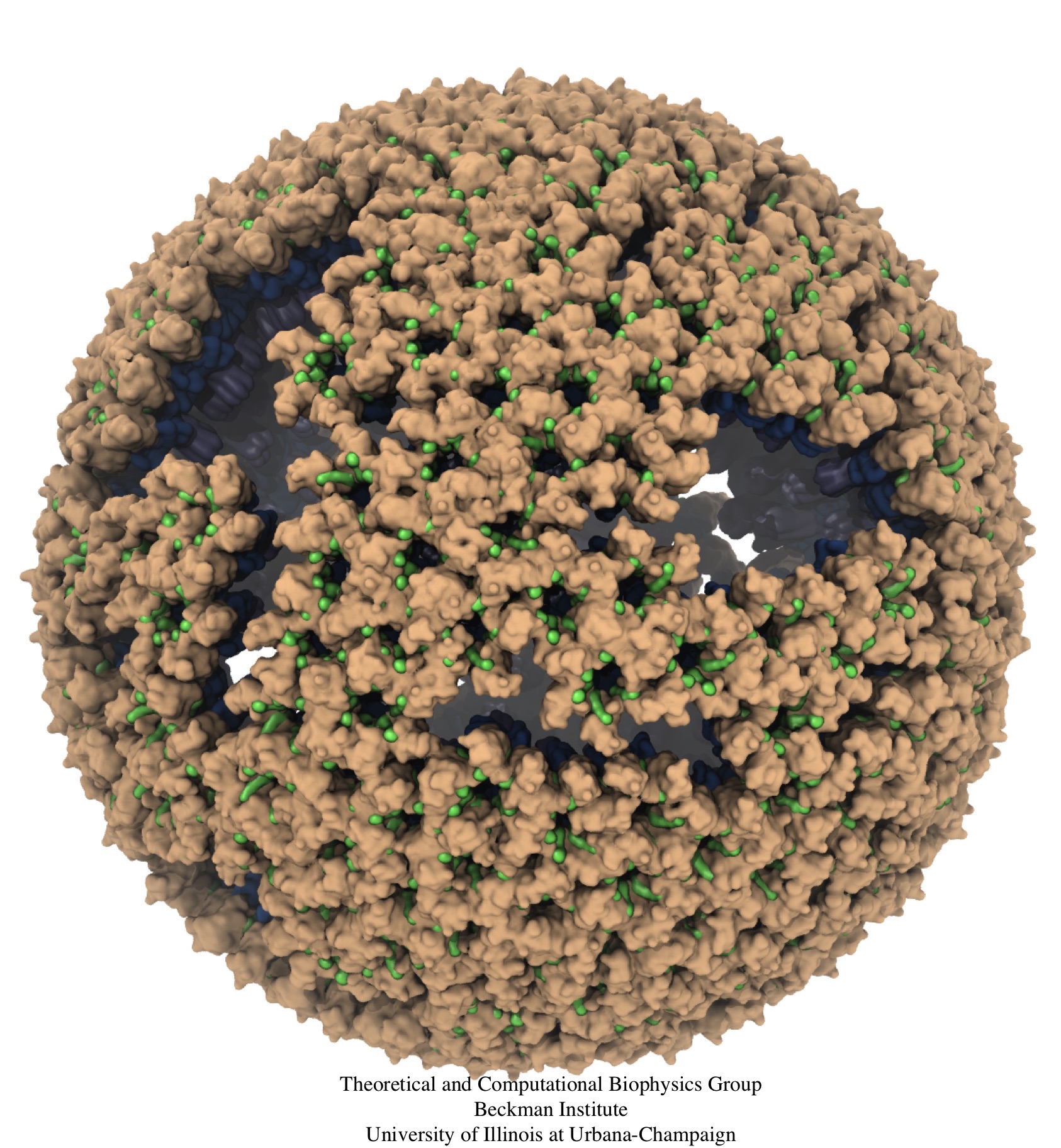
image size:
2.6MB
made with VMD
Retroviruses are parasites that pose a major health threat to humans (for example in case of HIV) and other animals (for example in case of RSV, M-PMV, MLV, and many more viruses). After a retrovirus hijacks a cell, the infected cell produces multiple copies of the virus which are then released into the host's bloodstream. These newly released viruses must mature before they can infect other cells. A strategy for preventing virus spread is therefore, to lock the viral particles in their immature, non-infectious state. However, to render the immature virus an attractive target for structure-based drug development one needs to know its chemical structure. Unfortunately, the complexity and size of the viral particle ― an incomplete hexagonal shell with a size close to 100 nm ― have prevented the experimental determination of the chemical, namely atomic level, structure of the virus. As reported recently, a team of computational and experimental researchers have provided an atomic structure of the immature retroviral lattice for the Rous Sarcoma Virus. The multi-domain RSV model was derived through a combination of state-of-the-art modeling techniques, including, cryo-EM-guided homology modeling, large-scale molecular dynamics simulations using enhanced sampling capabilities available in NAMD, together with experimental measurements such as X-ray crystallography and a wealth of biochemical data. Particularly, the model reveals novel features of the packing and dynamics of the immature capsid protein with implications for the maturation process and confirms the stabilizing roles of the so-called upstream and downstream domains of the immature RSV. More information is available on our retrovirus website, and in a highlight video.
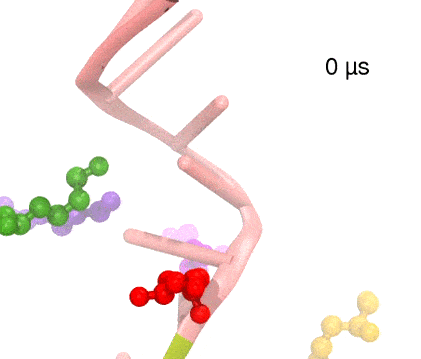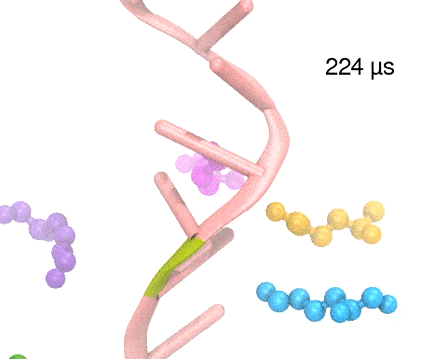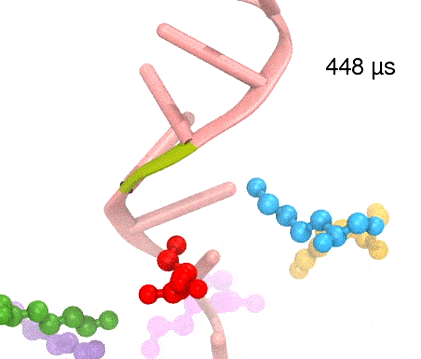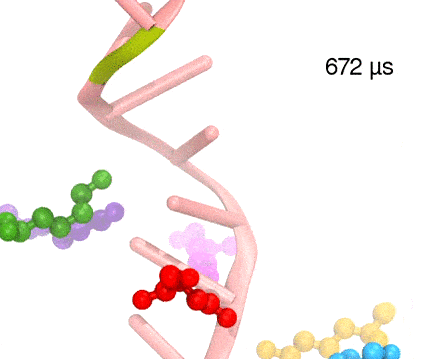
image size:
680.4KB
made with VMD
Many processes in living cells require molecular motors. Examples are transport of cargo within a cell, degrading misfolded proteins, and controlling gene expression. In the latter case acts a motor, called Rho, that moves along messenger RNA. The energy of the cell's motors stems from molecules of ATP that are converted to ADP, release thereby energy and drive motor action. How exactly this happens remained largely a mystery, despite decades of study and despite the availability of detailed molecular structures of the motors. A molecular dynamics study employing NAMD has achieved a great breakthrough in resolving the mechanism by which ATP-to-ADP conversion drives Rho to translocate along RNA. While molecular dynamics simulation, in principle, is well suited to explain Rho's motor action, the problem was that the action takes about a millisecond which is a time period beyond such simulations' reach. Employing new sampling methods, a recent publication, reported in new, complete and fascinating detail how Rho works. The simulations permitted literally to look under the hood of Rho's engine and see how it pulls itself along RNA and coordinates a cyclic and repeated motor action. It turned out that Rho, a ring of six identical protein subunits, engages in an ATP-to-ADP conversion-induced periodic motion of the subunits that pushes RNA electrostatically through the ring center. A completely surprising finding was the existence of coordination switches that make each ATP-to-ADP conversion lead to exactly one forward step along the RNA and keep the six subunits strictly synchronized, turning a randomly moving protein system into a well-behaved engine. More on our molecular motor website.
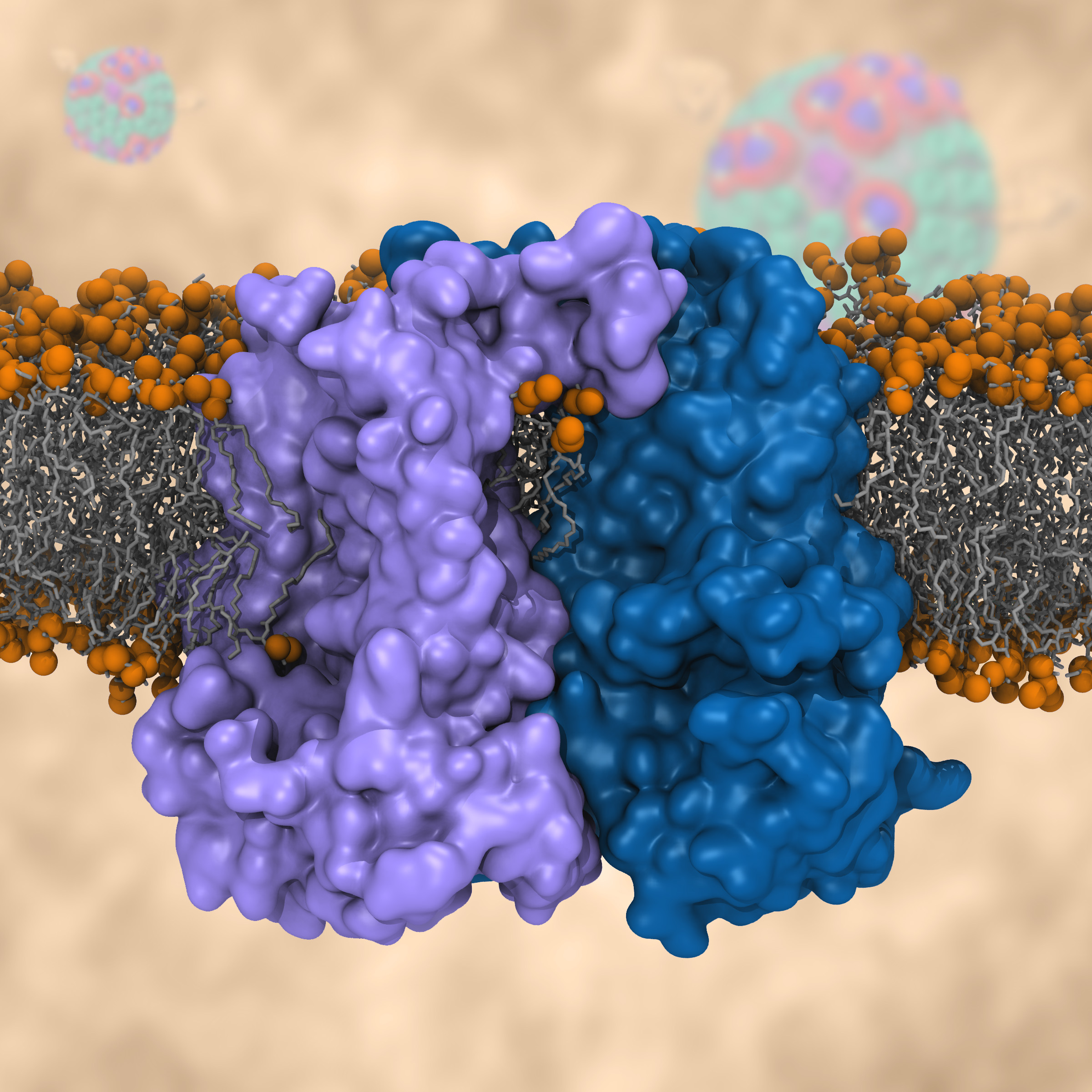
image size:
1.3MB
made with VMD
Most living cells acquire their energy through photosynthesis or respiration, both of which convert input energy (sun light or food, respectively) through coupled electron and proton transfer processes. A key role is played here by a protein, called the bc1 complex, that intermediately stores energy through the reaction of molecules of quinol into molecules of quinones, utilizing energy released to pump protons across an intracellular membrane. This reaction is initiated in the bc1 complex at the site of binding of the quinol molecule, but critical details about the physical mechanism leading to coupled electron-proton transfer are still unknown. A recent study, based on molecular modeling with NAMD and quantum chemistry calculations, investigated possible reaction mechanisms in case of the bc1 complex from the bacterium Rhodobacter capsulatus. The calculations suggest a novel configuration of amino acid residues responsible for quinol binding in the bc1 complex, and support a mechanism for coupled proton-electron transfer from quinol to iron-sulfur cluster. The study opens the door for a complete simulation description of the crucial role of the bc1 complex in bioenergetics. More about the bc1 complex can be found here.
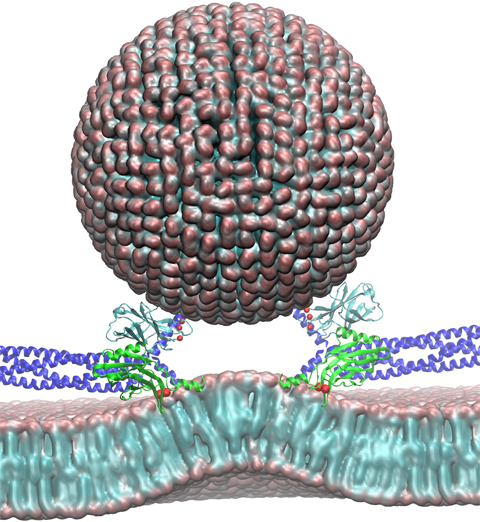
image size:
258.8KB
made with VMD
Neurons in the brain form a closely knit communication network with other neurons. Each neuron sends messages through up to thousands of cell-cell communication channels, so-called synapses. To avoid communication chaos, the messages of each neuron are chemically encoded as if neurons speak English to some neurons and French to others. The neurons employ an extremely efficient encoding system, packaging chemical message molecules, so-called neurotransmitters, in spherical vesicles encapsulated by a lipid membrane just like the whole neuron is encapsulated by a membrane. The vesicles aggregate near the presynaptic site of the membrane, ready to release their neurotransmitters into the space between neurons at the synapse, the so-called synaptic cleft. When a sender neuron becomes electrically active, as it wants to "speak", the electrical activity releases Calcium ions at the pre-synaptic cell that trigger merging (fusion) of vesicles with the sender neuron's membrane. At this point the neurotransmitter molecules flow into the synaptic cleft. The receiver neuron "hears" the signal by receiving the neurotransmitter molecules on receptors in the postsynaptic membrane, inducing as a result an electrical signal in the receiver neuron. The release involves a group of proteins that make vesicles ready for the release and proteins that execute the Calcium-triggered step, among the latter synaptotagmin I. As reported recently, researchers have proposed with the help of computer simulations using NAMD how synaptotagmin I acts. The simulations suggest that the experimentally observed structure of synaptotagmin I measured in vitro in a crystal/NMR form of the protein differs from the active, in situ form of synaptotagmin I. The finding, if true, will be a dramatic example for the role of computing in biology where the computer often complements observation studying, as in the present case, biomolecules in situ, namely their natural environment, rather than in vitro, namely in an artificial environment. Please read more on our neuron transmission website.
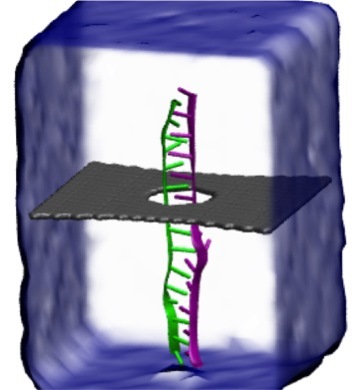
image size:
287.1KB
made with VMD
Threading DNA through a nanometer-size pore, so called nanopores, drilled into an ultrathin graphene membrane is a promising approach to build nanobiosensors for sequencing the human genome. Graphene nanopores can detect translocating DNA by recording concomitant flow of charged ions through the pore (see December 2011 highlight). As reported in the December 2013 highlight, graphene, which is an electrical conductor, offers a new way of sensing DNA molecules by monitoring sheet currents along the graphene membrane. DNA is a highly extensible molecule and upon mechanical manipulation can change its structure from a canonical helical conformation to a linear zipper-like conformation. A new study, which combines classical molecular dynamics simulations using NAMD with quantum mechanical simulations, suggests that sheet currents, in graphene membranes, can be used to detect conformation and sequence of a DNA molecule passing through the nanopore. This new research will guide the development of graphene-based nanosensors for DNA detection. More information can be found on our graphene nanopore website.
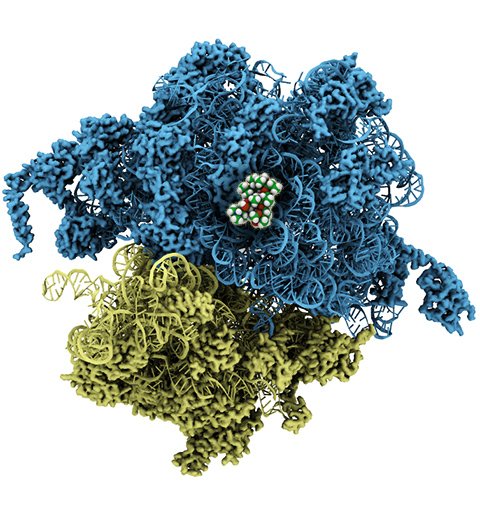
image size:
210.2KB
made with VMD
The ribosome, one of the ubiquitous molecular machines in living cells, is responsible for the critical task of translating the genetic code into functional proteins (See also Managing the Protein Assembly Line). The bacterial ribosome is the target of over 50% antibiotic drugs, for example, the clinically important macrolide family, including the widely-prescribed erythromycin (ERY) which is on the WHO essential medicines list. The antibiotic action of macrolide drugs has been known for over 50 years, however, the molecular mechanisms underlying the effects of these drugs are still unknown. It was previously believed that the antibiotic action by macrolide drugs has to be assisted by the presence of a nascent protein inside the ribosome. However, in a recent study, computational investigations jointly with biochemical experiments have revealed that the macrolide drugs can take an antibiotic action by altering the structure of the bacterial ribosome before translation of nascent protein really begins. Please see more highlights on translational control of the ribosome: Born to Control, Shutting Down the Protein Factory. Read more on our Ribosome website.



