Research Projects
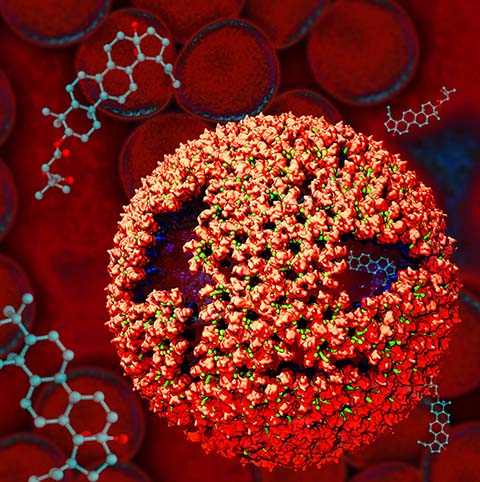
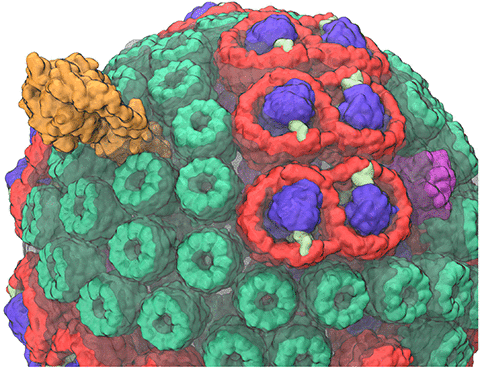
Viruses are small intracellular parasites that invade the cells of virtually all known organisms. They reproduce by utilizing the cell's machinery to replicate viral proteins and genomic material, generally damaging or killing the host cell in the process; subsequentelly, a large number of newly generated viruses go on to infect other cells. Viruses are responsible for a wide variety of human diseases, ranging from the common (influenza and colds) to the exotic (AIDS and West Nile virus, and Zika). Some viruses which are not dangerous to humans can also be exploited in technological applications, in addition, viruses find use in genetic engineering applications and increasingly in the design of new nanomaterials. At the very least, all viruses contain two components: the capsid (a protein shell), and a genome, consisting of either DNA or RNA. Some viruses also include accessory proteins to aid in infection, and in some cases a lipid bilayer to further protect their contents from the environment. The viral life cycle itself is deceivingly simple: viruses enter the cell, typically (but not always) through the interaction of their capsid with a receptor on the cell surface; the virus must then somehow disassemble its capsid to release its genetic material and any necessary helper proteins. The viral genome is then replicated and the proteins it codes for are synthesized to produce the raw material for the production of new viral particles; these new viruses then assemble and bud from the cell either through the membrane or upon cell death.
Bacterial communities within the human body greatly influence human health and play a significant role in disease predisposition, pathogenic, physical fitness, and dietary responsiveness. Importantly, bacteria utilize highly cooperative macromolecular machines to accomplish many cellular functions. Here we seek to understand, with molecular and atomistic fidelity, two such machines: the cellulosome and the chemosensory array, which underlie the phenomena of bacterial plant fiber degradation and chemotaxis respectively.
Biofuels: Bacteria can make a living off a very wide range of food sources. This agnosticism enables them to, among other things, serve as essential symbionts in animal digestive tracts where they assist their hosts in degrading cellulose fibers into metabolizable compounds. In particular, bacteria in the rumen of the cow face an especially tough job (see Tight Job in the Gut), digesting the hardy cellulose fibers of grasses. Key to their task are molecular tentacles on the cell surface of certain gut bacteria, so-called cellulosomes (pictured right), which develop a tight grasp on cellulose and then effectively cleave the molecules. In general, human gut bacteria (and their role in the broader human microbiome) are one of the most intensely researched topics in medicine.
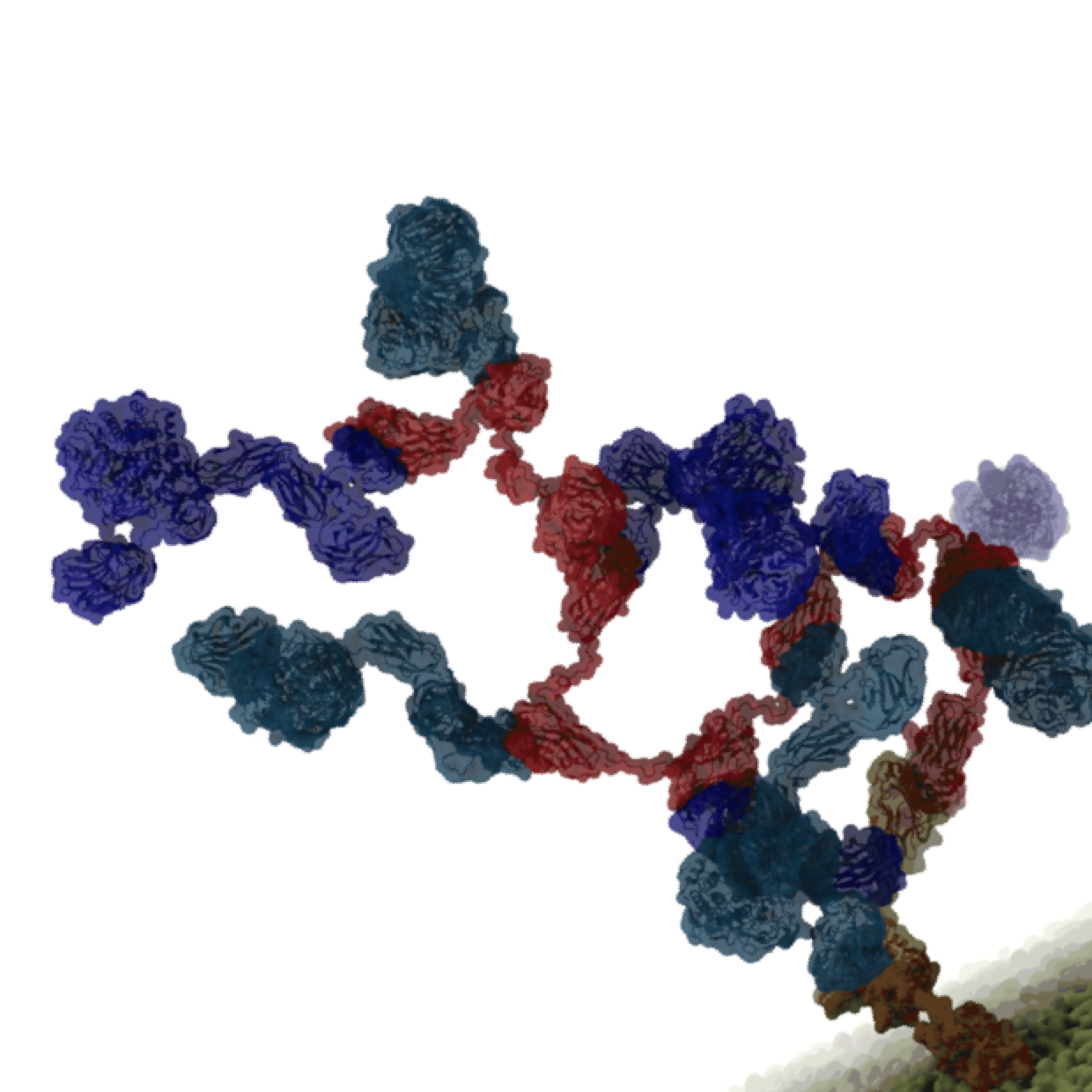
Bacterial Chemotaxis: Bacteria monitor their environments and respond by way of a fundamental sensory capability known as chemotaxis---one of the best studied behavioral systems in biology. Chemotactic responses in bacteria involve large complexes of sensory proteins, known as chemosensory arrays, that process the information obtained from the bacteria's habitat to determine its swimming pattern. In this sense, the chemosensory array functions as a bacterial brain, transforming sensory input into motile output. Despite great strides in the understanding of how the chemosensory array's constituent proteins fit and work together, a high-resolution description has, until recently, remained elusive (see Computing the Bacterial Brain). Here we are combining computational and experimental techniques to explore in detail the molecular mechanisms underlying sensory signal transduction and amplification within this amazing biological apparatus.
Many cellular processes are driven by molecular motors, specialized proteins that utilize the energy generated from chemical reactions to perform physical work. Molecular motors play key roles in, for example, muscle contraction, protein degradation and recycling, cargo transport, and cell motility. Defects in motor function are implicated broadly in cancer, as well as numerous cardiovascular, neurological, and reproductive diseases. Researchers in the Theoretical and Computational Biophysics Group are interested in studying the complex conformational transitions that underlie the chemo-mechanical action of molecular motors toward characterizing their mechanisms and relationships to human disease.
Biosphere is powered directly or indirectly by harvesting the energy of sunlight. Plants and bacteria convert solar energy into chemical energy, which is used, ultimately, for producing food. Compared to the complicated energy harvesting apparatus of plants, the primitive purple bacteria display a far simpler instance of photosynthesis. In purple bacteria, the energy harvesting processes are performed by a spherical vesicle, called the chromatophore, featuring hundreds of cooperating proteins assembled together on a membrane. The energy conversion process in the chromatophore shows that the bacteria adapted to efficiently harvest light at the dim light conditions typical of their habitat. (see Bacteria Harvest Light Efficiently).
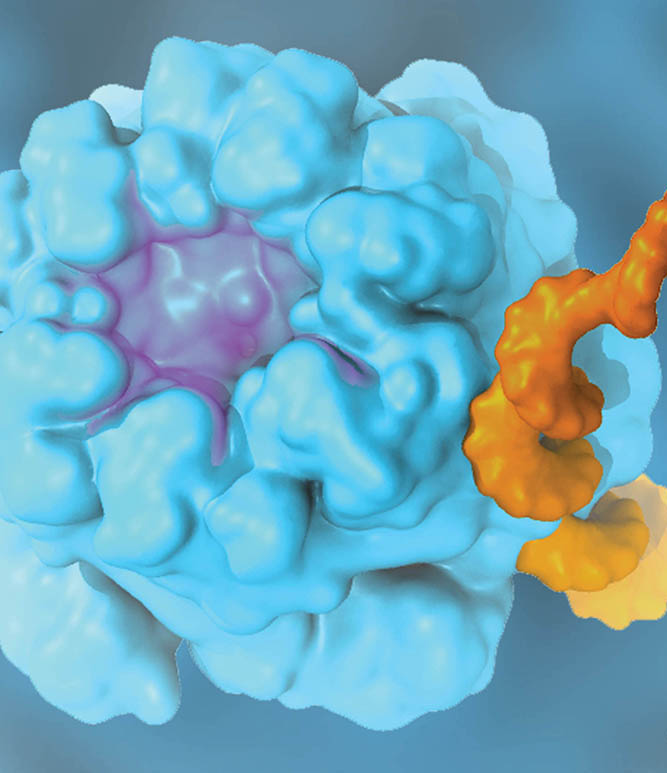
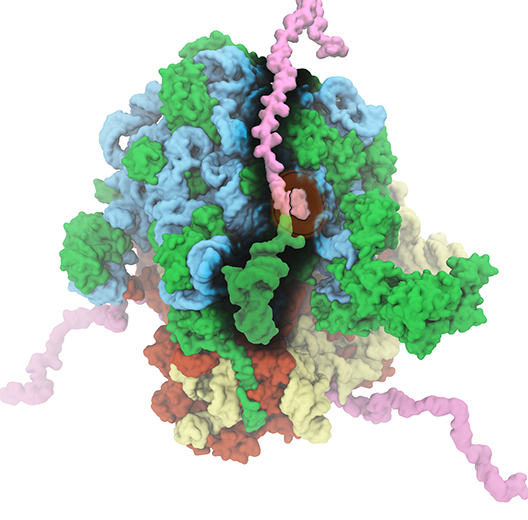
The translation of genetic information into proteins is essential for life. At the core of this process lies the ribosome, a quintessential large (2.5-4.5 MDa) molecular machine responsible for translating genetic material into functional proteins. In a growing cell, ribosomes comprise up to half of the net dry weight. Because of its fundamental role in the cell, 50% of all efforts to develop antibiotics target bacterial ribosomes, taking advantage of the structural differences between bacterial and human ribosomes. The Theoretical and Computational Biophysics Group has a long-standing history of investigating the structure and function of ribosome. In collaboration with leading experimental scientists, TCBG researchers have also studied several ancillary proteins, which through their interplay with ribosome facilitate the process of protein synthesis and protein installment in the case of membrane proteins. Elongation factors EF-Tu and EF-G as well as membrane protein insertases such as SecYEG and YidC are examples of these studied ribosomal ancillary proteins.
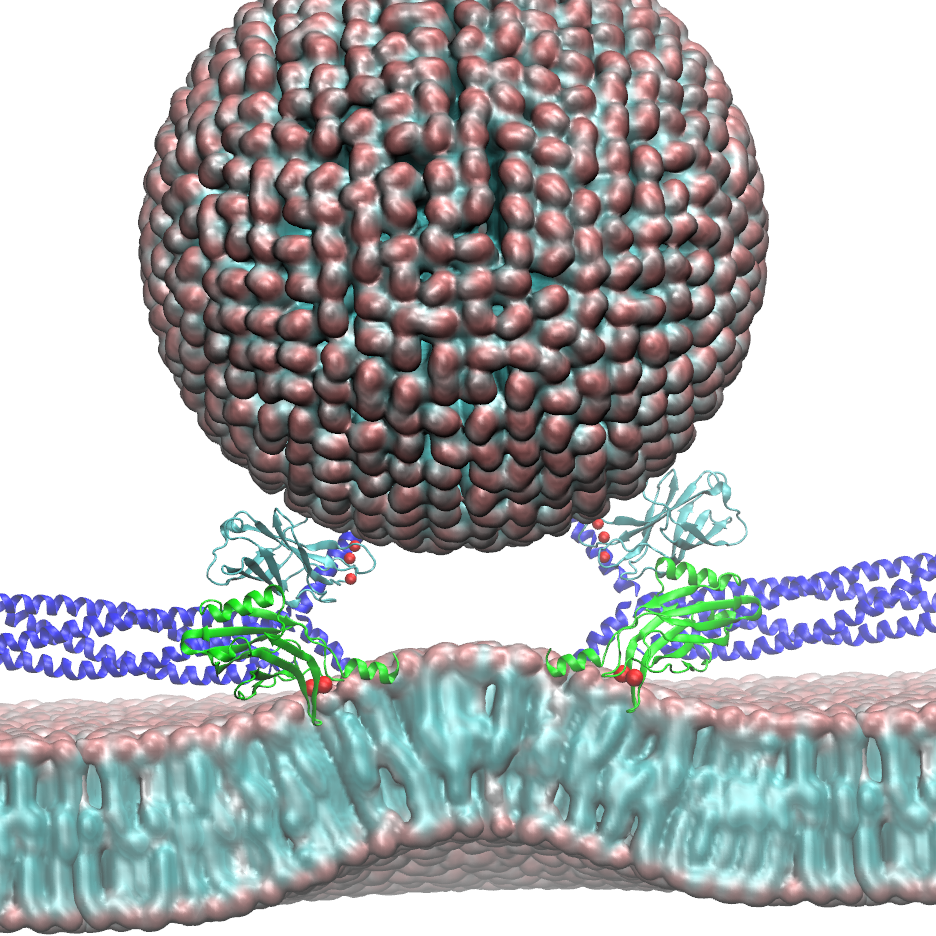
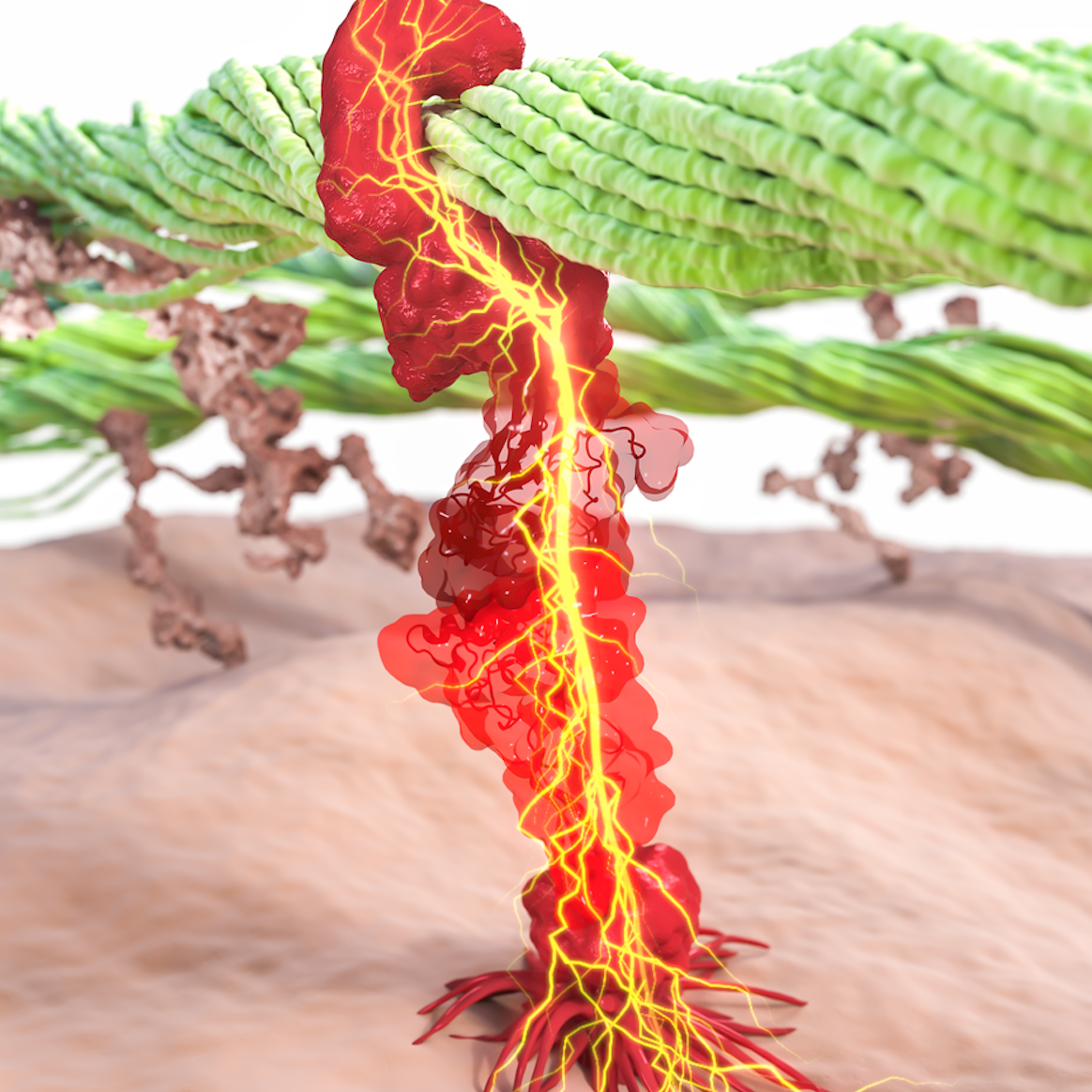
Cells explore their environment by sensing and responding to mechanical forces. Many fundamental cellular processes, such as cell migration, differentiation, and homeostasis, take advantage of this sensing mechanism. At molecular level mechanosensing is mainly driven by mechanically active proteins. These proteins are able to sense and respond to forces by, e.g., undergoing conformational changes, exposing cryptic binding sites, or even by becoming more tightly bound to one another. In humans, defective responses to forces are known to cause a plethora of pathological conditions, including cardiac failure, pulmonary injury and are also linked to cancer. Microorganisms also take advantage of mechano-active proteins and proteins complexes. Employing single-molecule force spectroscopy with an atomic force microscope (AFM) and steered molecular dynamics (SMD) simulations we have investigated force propagation pathways through a mechanically active protein complexes.
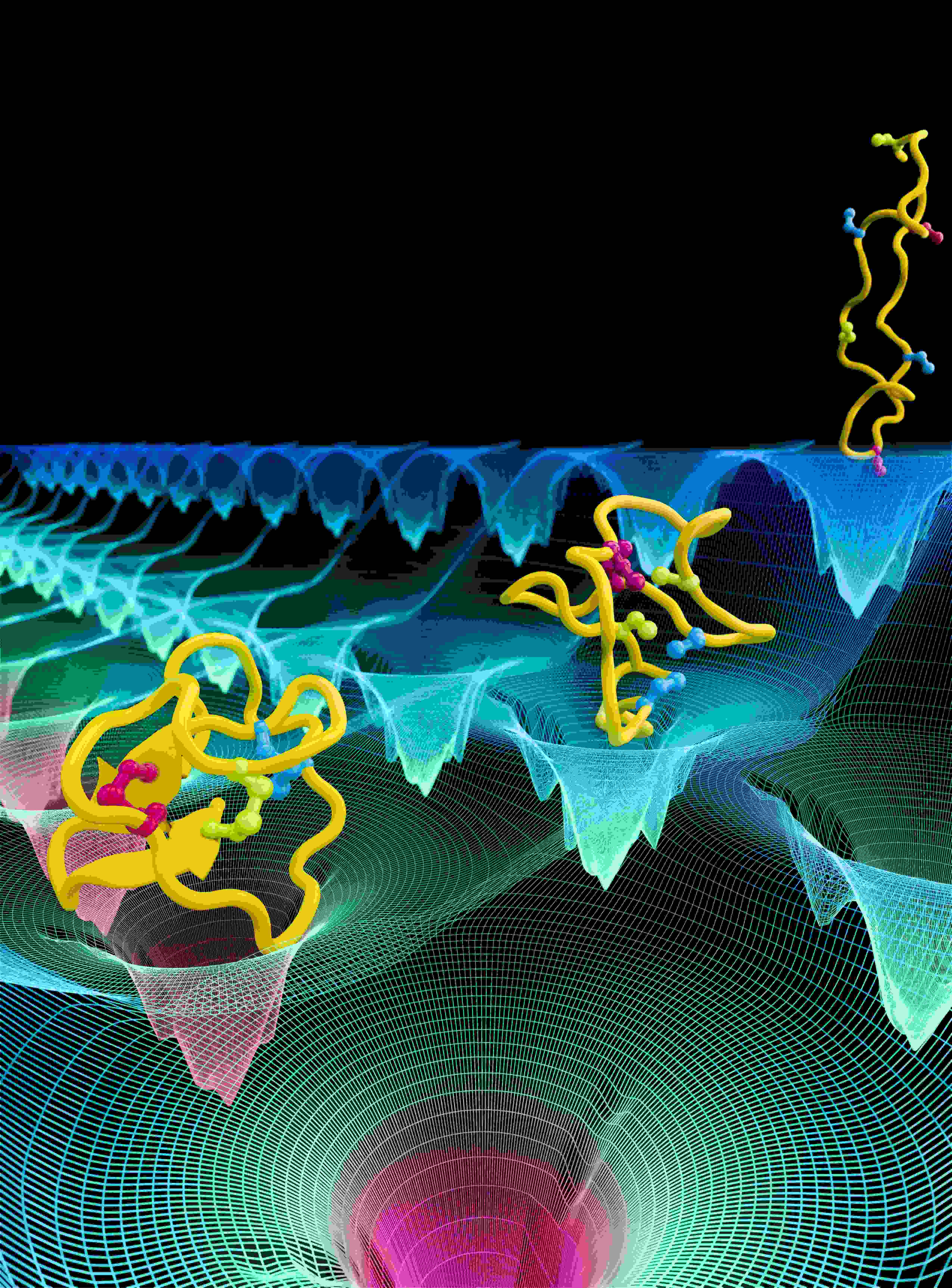
Many structures of biological molecules determined today are created by combining many disparate sources of experimental data. Techniques such as cryo-electron microscopy (cryo-EM), X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and small-angle X-ray scattering (SAXS), each provide unique and powerful means for characterizing critical aspects of a molecular structure. However, no single experiment can produce a comprehensive all-atom model for many biomolecules, especially those that are too large or complex. For example, X-ray crystallography furnishes atomic-level detail of a molecule, though the results may not necessarily capture the system in a physiologically-relevant state. On the other hand, cryo-EM studies produce low-resolution maps depicting the system in native-state conformations. Computation can now be used to integrate such data coming from different sources in order to produce a model that matches best to all available data.