Molecular Motors
Many cellular processes are driven by molecular motors, specialized proteins that utilize the energy generated from chemical reactions to perform physical work. Molecular motors play key roles in, for example, muscle contraction, protein degradation and recycling, cargo transport, and cell motility. Defects in motor function are implicated broadly in cancer, as well as numerous cardiovascular, neurological, and reproductive diseases. Researchers in the Theoretical and Computational Biophysics Group are interested in studying the complex conformational transitions that underlie the chemo-mechanical action of molecular motors toward characterizing their mechanisms and relationships to human disease.
Spotlight: RNA Shredder (May 2016)
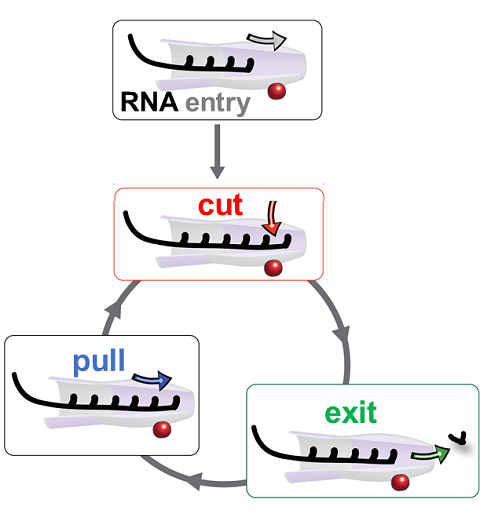
image size:
386.7KB
made with VMD
RNA molecules are continuously synthesized in living cells as carriers of biological information written in the sequence of basic RNA units, called nucleotides. To keep cells healthy, RNA molecules not longer needed or with errors have to be removed. A large barrel-like protein complex, the RNA exosome, is a molecular machine that degrades unneeded RNA molecules, pulling them inside its long internal channel and cutting them sequentially into single nucleotides. A new molecular dynamics study, employing NAMD, shows that a special active protein subunit of the exosome, called Rrp44, grips tightly the RNA molecule throughout its extended channel. Rrp44 grips RNA molecules with five or more nucleotides in length while their ending nucleotides get sequentially cut, whereas shorter RNAs are only weakly bound and unlikely to be cut. The simulations reveal how the exosome can act both as a molecular motor that pulls RNA, without energy input other than the one released in nucleotide cleavage, as well as an enzyme that cuts RNA. More information is available on our RNA exosome website.



