NAMD, recipient of a 2002 Gordon Bell Award, a 2012 Sidney Fernbach Award, and a 2020 Gordon Bell Prize, is a parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. Based on Charm++ parallel objects, NAMD scales to hundreds of cores for typical simulations and beyond 500,000 cores for the largest simulations. NAMD uses the popular molecular graphics program VMD for simulation setup and trajectory analysis, but is also file-compatible with AMBER, CHARMM, and X-PLOR. NAMD is distributed free of charge with source code. You can build NAMD yourself or download binaries for a wide variety of platforms. Our tutorials show you how to use NAMD and VMD for biomolecular modeling.
Breaking News
| NAMD 3.0 Release - Stable release featuring new GPU-resident mode for NVIDIA and compatible AMD GPUs. Updates to Colvars, Charm++, and user guide documentation. |
| NAMD 3.0 New Features - webpage is posted |
| NAMD GPU-resident benchmarks - results and data sets with GPU-optimized configuration posted |
| NAMD 2.15 ALPHA Release providing GPU-offload support for Intel GPU Max Series. This source code release available on the download page includes SYCL code that can be built using the Intel oneAPI toolkits. Following the download link reveals a page with detailed build instructions. |
Spotlight: Sticky Bugs (May 2018)
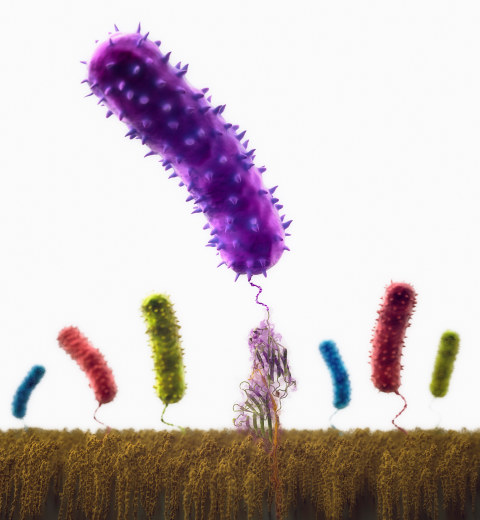
Staph infections are caused by bacteria commonly found on the skin of healthy individuals, where they can only cause relatively minor skin problems. However, when staph bacteria enter a person's bloodstream, they can travel to locations deep within the body causing infections that are often hard to treat. Not surprisingly, staph infections are the leading cause of healthcare-related, so called nosocomial infections. Particularly vulnerable areas are medical devices such as artificial joints or cardiac pacemakers, where the bacteria strongly stick to through formation of biofilms. Central to biofilm formation is an unusually tight interaction between microbial surface proteins called adhesins and the extracellular components of the host cells. In a recent report in Science , researchers used a combination of atomic force microscopy and GPU-accelerated molecular dynamics simulations using QwikMD and NAMD , to explore how the connection between adhesin and its target fibrinogen peptide can withstand huge forces greater than 2 nano-Newtons (see also Perspective in Science. ). The geometry and molecular details of the interaction ensures that, when pulled, the load is distributed over many hydrogen bonds all of which need to be broken at once before separating the bacterium from the surface of the host cell (see video on YouTube ). The unexpected, new mechanism, expands our understanding of why pathogen adhesion is so resilient and open new avenues for an intelligent design of antimicrobial therapies through development of anti-adhesion drugs. Read more in Science.
Overview
Why NAMD? (in pictures)
How to Cite NAMD
Features and Capabilities
Performance Benchmarks
Publications and
Citations
Credits and Development Team
Availability
Read the License
Download NAMD Binaries
(also VMD)
Build from Source Code
- Git access now available
Run at NCSA, SDSC, NICS, or Texas
Training
NAMD Developer Workshop in Urbana (August 19-20, 2019)
PRACE School on HPC for Life Sciences (June 10-13, 2019)
"Hands-On" Workshop in Pittsburgh (May 13-17, 2019)
Charm++ Workshop in Urbana (May 1-2, 2019)
Enhanced Sampling and Free-Energy Workshop (Sept 10-14, 2018)
NAMD Developer Workshop in Urbana (June 11-12, 2018)
"Hands-On" Workshop in Pittsburgh (May 21-25, 2018)
"Hands-On" QM/MM Simulation Workshop (April 5-7, 2018)
Older "Hands-On" Workshops
Support
Mailing List Issues for Yahoo.com Addresses
Announcements
NAMD 3.0 Release (June 2024)
NAMD 3.0 New Features (Feb 2024)

NAMD 2.14 Bug Fixes (Apr 2022)
NAMD 2.14 Release (Aug 2020)
NAMD 2.14 New Features
One-click NAMD/VMD in the cloud
QM/MM Interface to MOPAC and ORCA
QwikMD GUI Released in VMD 1.9.3
Previous Announcements
Documentation
Related Codes, Scripts, and Examples
NAMD Wiki (Recent Changes)
Older Documentation
News
Sparing healthy microbes while using a novel antibiotic

AMBER force field use in NAMD for large scale simulation

NAMD GPU-resident benchmarks available

NAMD and VMD share in COVID-19 Gordon Bell Special Prize
NAMD reference paper published online
Coronavirus Simulations by U. Delaware Team
Coronavirus Simulations on Frontera Supercomputer
Breakthrough Flu Simulations
Oak Ridge Exascale Readiness Program
Prepping for Next-Generation Cray at NERSC
Supercomputing HIV-1 Replication
How GPUs help in the fight against staph infections
Computational Microscope Gets Subatomic Resolution
Opening New Frontiers in the Battle Against HIV/AIDS
HIV Capsid Interacting with Environment
Assembling Life's Molecular Motor
Older News Items