Highlights of our Work
2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
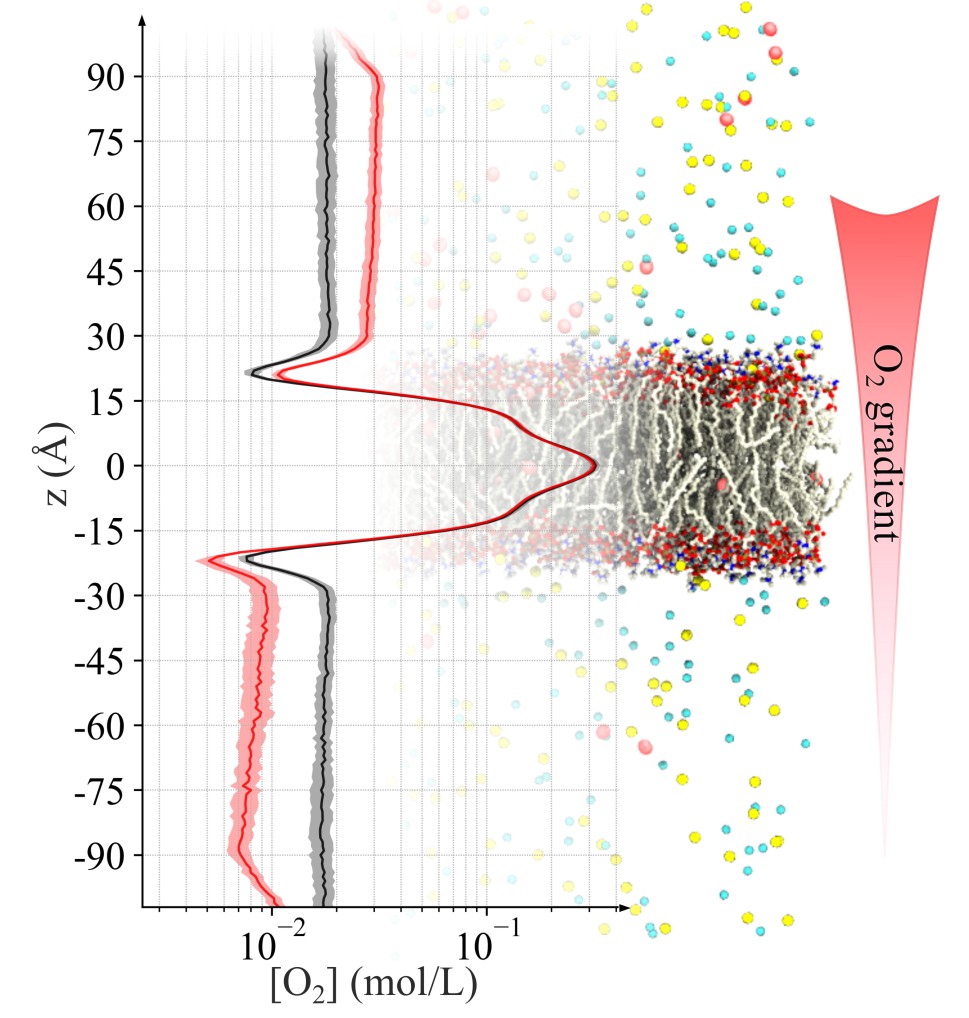
Permeation of metabolic substrates across biological membranes is a fundamental process in cellular life. This process is largely driven by the concentration gradient of various molecules between the outside and inside of a cell. To meet the need for creating such concentration gradients in MD simulation, and to calculate permeation under natural conditions, we developed a technique in NAMD to continually drive permeant molecules near the periphery of the simulation box across the periodic boundary, which results in a sustained gradient in the center of the simulation system where the membrane is located. This allows for purely diffusive motion of particles across a membrane, enabling one to directly calculate permeability the same way as in experiment. Read more in a recent paper.