Light Harvesting Complex II
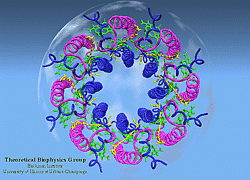 |
LH-II of Rs. molischianum
(figure generated with the program VMD) |
The photosynthetic apparatus of purple bacteria consists of two types of pigment-protein complexes: the reaction centers and the light-harvesting complexes. The main function of the light-harvesting complexes is to gather light energy and to transfer this energy to the reaction centers for the photo-induced redox processes. In most purple bacteria, the photosynthetic membranes contain two types of light-harvesting complexes: light harvesting complex I (LH-I) and light harvesting complex II (LH-II) [1]. While LH-I is tightly bound to the photosynthetic reaction centers, LH-II is not directly associated with the reaction centers, but transfers energy to the reaction centers via LH-I [1].
Studies of primary charge separation are well advanced [2] ever since the discovery of the three-dimensional structure of the photosynthetic reaction center by Michel et al [3]. However, our knowledge of energy transfer is lagging behind due to lack of structural information about light-harvesting complexes. It was known that energy transfer from light harvesting complexes to the RC occurs within 100 PS and with high efficiency (near 100%) [4]. Little was know about the pathways of energy transfer and the underlying energy transfer mechanisms. Unlike electron transfer reactions which occurs locally at the RC, energy migration from light harvesting complexes to the RC is a global event, requiring concerted actions, either sequential or parallel, of all the pigment protein complexes including LH-IIs, LH-I and the RC. To understand the mechanism for energy transfer within the photosynthetic system, structural information of each component and of the overall assembly of all the components is needed.
In the past two years, major breakthroughs in the forefront of bacterial pigment-protein structures have changed prospect radically. High resolution crystal structures of LH-II complexes from two species (Rps. acidophila and Rs. molischianum) have been determined [5,6]. We have determined the structure of the Light-Harvesting Complex II of Rs. molischianum in collaboration with H. Michel [6]. The LH-II complex of Rs. molischianum had been crystallized by Michel, and X-ray diffraction data had been collected up to 2.4 Angstrom resolution. Resolving a structure from measured diffraction intensities requires knowledge of phases which is unobtainable from a single diffraction experiment. Conventionally, the phase problem is solved by means of the multiple isomorphous replacement method [7] which, in the present case, has proven unsuccessful due to technical difficulties in making heavy metal derivatives. An alternative solution to the phase problem is to phase the structure by using a homologous structure in a procedure called molecular replacement [8]. In this method, a homologous probe structure is fit into the unit cell of the crystal and used to generate initial scattering phases. When this project was initiated, there existed no structure homologous to LH-II of Rs. molischianum. Our solution to the problem was to computationally model a structure for LH-II of Rs. molischianum [9,10] and to utilize the modeled structure in the framework of molecular replacement method to phase the X-ray diffraction data [6].
Most recently, we have computationally modeled an atomic structure for LH-I of Rb. sphaeroides based on a high degree of homology of the alpha-beta heterodimer of LH-I from Rb. sphaeroides to that of LH-II of Rs. molischianum. An LH-I - RC complex was subsequently formed by energy minimization under constraints of biochemical data. Thus, for the first time, we have achieved a complete picture of pigment organization in the PSU at atomic resolution. This achievement lays the foundation for the studies on energy transfer mechanisms in the bacterial photosynthetic membrane.

|

|
Three-dimensional hardcopy of the light harvesting system LH-II as shown above. The model is built together from eight heterodimer units which were produced at the telemanufacturing facility of the San Diego Supercomputer Center in collaboration with M. Bailey. Both figures show the complete, circular aggregate. Blue: alpha apoprotein; red: beta apoprotein; green: bacteriochlorophyls; yellow: lycopenes.



