Mechanical Proteins
Mechanical force is ubiquitous in biological molecular processes, such as in stretching cytoskeleton inside cells, in assembling fibrils connecting cells, and in regulating ligand-receptor association at the cell surface. Correspondingly, there are numerous proteins that participate in such processes involving mechanical forces. We term these proteins mechanical proteins. Although the folded structure of many mechanical proteins in equilibrium states has been resolved, little is known about the dynamic response of these proteins when subjected to mechanical forces in cell environment. To bridge this gap, we have simulated the non-equilibrium transformations of mechanical proteins using steered molecular dynamics. Here we describe the mechanical properties of the extracellular matrix (ECM) protein fibronectin and the muscle protein titin.
The Mechanical Functions of Fibronectins in the Extracellular Matrix
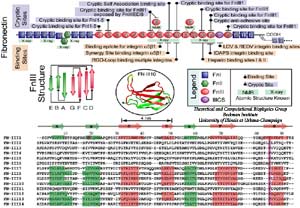
| Mechanical stress has been found to regulate the assembly of FN fibrils, a process termed fibrillogenesis, through integrin receptors that mechanically couple intracellular actin filaments to extracellular FN molecules. It has been hypothesized that the stretching of FN fibrils unfolds individual FN-III modules, providing the necessary extension to the FN fibril that can be stretched four times as long as its relaxed length. In addition to providing elasticity, the unfolding of FN-III modules is functionally relevant to mediating fibrillogenesis by exposing otherwise buried cryptic sites. |
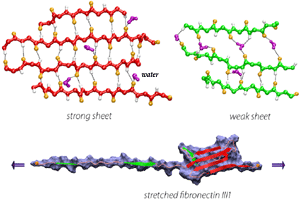
In collaborating with research groups (Viola Vogel and David Craig, Univ. of Washington; Olivier Lequin, Pierre & Marie Curie University; Iain Campbell, University of Oxford), we have combined NMR and computer modeling to study the 3D atomic structures and mechanical response of fibronectin type III1 (FN-III1) module (Gao et al. PNAS 2003). Like other FN-III modules, FN-III1 exhibits two anti-paralleled beta sheets packed into the beta-sandwich motif. However, due to the substitutions of two key residues (one located at B-strand, and the other located at G-strand) compared to other FN-III modules, FN-III1 is designed by nature so that one beta-sheet is stronger than the other one. Once subjected to tensile force generated by cells, FN-III1 unravels the weaker beta-sheet first, exposing hydrophobic residues on the stronger sheet. These hydrophobic residues can bind to other FN modules, promoting the assembly of fibronectin fibrils. The partially unfolded FN-III1 with the stronger beta-sheet, also termed anastellin, can inhibit the growth of cancer cells. |
Movies of stretching FN-III1
|
 Unfolding pathways of FN-III10 |
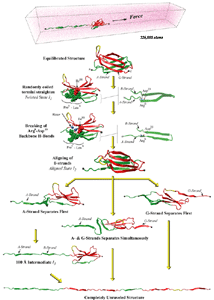
SMD simulations revealed that the main force-bearing elements of FN-III modules arise primarily from the beta-strands close to the termini, specifically, from hydrogen bonding and electrostatic packing between A- and B-strands and between F- and G-strands. FN-III modules may exhibit multiple unfolding pathways and intermediates as revealed in SMD simulations of the tenth FN-III module, FN-III10 (Gao et al., JMB, 2002). The simulations were carried out in water boxes that allow up to full extension of a single FN-III10 module. Two intermediates, termed twisted and aligned states and are universal to other FN-III modules studied (Craig et al., PNAS, 2001; Craig et al., Structure, 2004), exhibit within short extension up to 70% of their original length without unraveling the tertiary structure. The aligned intermediate thus may expose partially buried cryptic binding sites. From the aligned state, FN-III10 progresses along three alternative unfolding pathways: separating first the A-strand from the module, separating the A- and G-strands simultaneously, or separating first the G-strand. When unraveling of the beta-strands began with the A-strand, a stable intermediate at 100 Å was observed. With four-fold extension compared to folded FN-III10, the intermediate may serve as a basis for fibronectin self-assembly through a proposed beta-strand swapping mechanism.
|
FN-III10 mediates cell adhesion to the ECM via its integrin binding motif, Arg-Gly-Asp (RGD), that is located at the apex of the loop connecting beta-strands F and G. The binding of the RGD loop to certain integrins is assisted by a synergy site on the neighboring domain FN-III9. SMD simulations found that this interaction can be regulated either through directly detaching the RGD motif (Krammer et al. PNAS, 1999; Gao et al., JMB, 2002) or through increasing the distance between the synergy site and the RGD loop (Krammer et, al, MB, 2002). By separating the G-strand from the remaining fold, the distance between the apex of the RGD-containing loop and the module surface was shortened. This conformational change reduces the affinity to integrin. The RGD loop thus constitutes a mechanosensitive switch for recognition by integrin receptors. SMD simulations of the FN-III9-10 dimer identified an intermediate in which the length of the linker chain between the two modules is increased by 17 Å, a change that could also switch off the signaling between FN modules and transmembrane protein integrins.
|
Movie of stretching FN-III10 |
 Mechanical stability of FN-III modules |
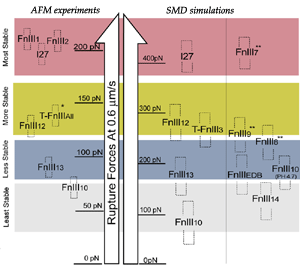
Despite remarkably similar tertiary structures, FnIII modules share low sequence homology. By contrast, the sequence homology for the same FnIII module across multiple species is notably higher, suggesting that sequence variability is functionally significant. Systematic SMD simulations have been conducted with on nine structurally known FN-III modules (Craig et al., PNAS, 2001; Craig et al., Structure, 2004). The simulations suggest that the mechanical stability of FnIII modules can be tuned through substitutions of just a few key amino acids by altering access of water molecules to hydrogen bonds that break early in the unfolding pathway. The hierarchy of mechaincal stability compared in SMD simulations are qualitatively in well agreement with atomic force microcopy (AFM) experiments (Oberhauser et al., Nature 1998, JMB 2002). Determination of such mechanical hierarchy in which FnIII modules unfold is essential to learn how cells use force to regulate the exposure of molecular recognition sites by stretching extracellular matrix proteins.
|
The Mechanical Functions of Immunoglobulin Domains in the Muscle Protein Titin
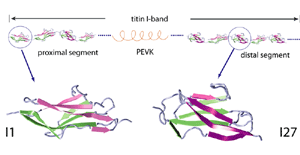
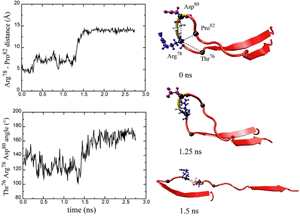
One important physiological function of titin is to develop a passive force which keeps sarcomere components uniformly organized during muscle contraction and restores sarcomere length when the muscle is relaxed. The I-band part of titin is believed to be responsible for this function. It consists mainly of two tandem regions of Ig domains (proximal Ig region and distal Ig region), separated by the N2 and PEVK regions. Each of the Ig domains possesses about 100 amino acids forming a beta-sandwich motif. The unique sequence regions like PEVK, however, mostly contains coiled conformations which are easily elongated when the muscle is stretched. When stretched beyond its resting length, titin I-band first extends links between domains from a coiled confirmation. Further stress results in the unfolding of the I-band polypeptide. The PEVK region unfolds first, followed by the unravelling of the N2 region. Unfolding of individual Ig domains occurs if additional extension is required. |
Movie of mechanical unfolding titin I27 |
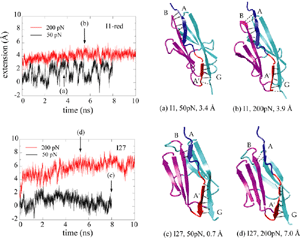
The reversible unfolding of titin Ig domains has been demonstrated in single molecule studies using AFM and optical tweezers. The AFM experiments have shown a characteristic sawtooth pattern in force-extension profiles, which can be attributed to the subsequent unraveling of several Ig domains. In order to understand the mechanical unfolding of titin Ig domains at atomic level, SMD simulations were performed on titin domain I27 (Lu et al., BJ, 1998; CP, 1999; Marszalek et al., Nature 1999). The SMD simulations of I27 revealed that a two-step unfolding pathway corresponding to two sequential events of interstrand hydrogen bond rupture, in which two sets of hydrogen bonds connecting beta-strands A and B, and beta-strands A' and G, are broken. At forces around 100 pN the first set of hydrogen bonds near the N-terminus breaks with a concomitant 4 to 7 Å extension, in agreement with the extension-force profile recorded in AFM experiments; the second set of hydrogen bonds breaks at forces of above 200 pN and initiates the complete unfolding. |
Unlike I27, the Ig domain from distal segment of titin I-band, Ig domains from approximal segment, such as I1, exhibit a slightly different mechanical design. Crystal structure of I1 displays a strong connection (six inter-strand hydrogen bonds) between I1's A- and B-strands, while a slightly weaker connection (five inter-strand hydrogen bonds) betweeen A'- and G-strands. SMD simulations of titin I1 domain revealed that the major mechanical resistance of I1 is contributed to both the hydrogen bonds between A- and B-strands and between A'- and G-strands (Gao et al., BJ, 2002). Under a constant force of 200pN, two hydrogen bonds near N-terminus of I1 break first. However, the extension of I1 gained from this breakage is limited because there are four remaining intact hydrogen bonds betweeen A- and B-strands. As a result, I1 is unlikely to exhibit the intermediate observed in unfolding of I27. Recent AFM experiments of I1 and its mutants confirmed these observations (Li, et. al, Nature 2002; JMB 2003). |
|
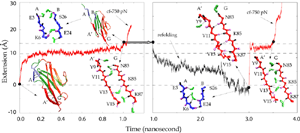 |
Water molecules participate actively the mechanical unfolding process, in particular, the breaking and re-forming of inter-strand hydrogen bonds that characterize the mechanical stability of titin Ig domains, as shown in detail analysis from SMD simulation trajectories (Lu et al., BJ, 2000). The refolding of the protein is a reversed process involving the driven out of water molecules by hydrophobic force. The reversible unfolding of I27 has been simulated (Gao et al., BJ, 2001). The equilibrated protein was initially stretched with 750 pN constant force until all hydrogen bonds between A- and B-strands and between A'- and G-strands are broken. The force was then released, and the partially unfolded I27 was subsequently allowed to spontaneously relax. During the 2 ns refolding simulation, the domain became more compact with its extension decreasing from over 15 Å to 0 Å, the extension of the native fold. At the end of the refolding simulation, five of the six hydrogen bonds connecting A' and G reformed by driving out water molecules. One of two A-B hydrogen bonds was also reformed. Re-applying a 750 pN constanct force generated a similar plateau that corresponds to breaking re-formed hydrogen bonds.
|
Publications
This page is created by Mu Gao, last updated 12/15/2003.
Click here to view the old titin page built by Hui Lu and Barry Isralewitz.