NAMD, recipient of a 2002 Gordon Bell Award, a 2012 Sidney Fernbach Award, and a 2020 Gordon Bell Prize, is a parallel molecular dynamics code designed for high-performance simulation of large biomolecular systems. Based on Charm++ parallel objects, NAMD scales to hundreds of cores for typical simulations and beyond 500,000 cores for the largest simulations. NAMD uses the popular molecular graphics program VMD for simulation setup and trajectory analysis, but is also file-compatible with AMBER, CHARMM, and X-PLOR. NAMD is distributed free of charge with source code. You can build NAMD yourself or download binaries for a wide variety of platforms. Our tutorials show you how to use NAMD and VMD for biomolecular modeling.
Breaking News
|
NAMD 3.0.2 Release -
Point release 3.0.2 fixes bugs in the Colvars module that affect not only NAMD 3.0 and 3.0.1, but also earlier NAMD 2 versions. See announce.txt for details regarding correctness issues in: eigenvector, orientationAngle, orientation, both coordNum and selfCoordNum with pairlist, and distanceZ with dynamic axis. For more information about the bugs in Colvars, please see Colvars Bugfix Announcement. We encourage all NAMD users to upgrade to version 3.0.2. |
| NAMD 3.0 New Features - webpage is posted |
| NAMD GPU-resident benchmarks - results and data sets with GPU-optimized configuration posted |
| NAMD 2.15 ALPHA Release providing GPU-offload support for Intel GPU Max Series. This source code release available on the download page includes SYCL code that can be built using the Intel oneAPI toolkits. Following the download link reveals a page with detailed build instructions. |
Spotlight: Bringing Oxygen into an Enzyme (Aug 2007)
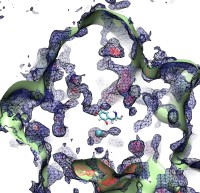
image size:
133.1KB
made with VMD
Because oxygen gas is very reactive, it is frequently employed by the cell as a reagent by proteins called enzymes, which build the organic compounds that the cell needs. One such enzyme belongs to the copper amine oxidase family. These proteins transform amine-containing compounds into molecules needed by the cell, by reacting the compounds with oxygen. Researchers have long been interested in finding out how the various reagents reach the buried copper active site before the final oxidation reaction can occur. While copper amine oxidases exhibit an obvious channel for capturing the amine compounds to be modified, it had been unclear until now how oxygen molecules make their way through the enzyme. With the help of computer simulations using NAMD, researchers have identified in a recent publication, the routes taken by oxygen inside various copper amine oxidases from different species. In order to accomplish this, they analyzed simulations of the motions of four copper amine oxidases, using the VMD analysis and visualization software, which can predict the probability of finding oxygen molecules anywhere inside the simulated proteins. This analysis found numerous oxygen conduction routes inside each copper amine oxidase, i.e., oxygen can enter the protein through many routes, as it would in a sponge. More information on finding O2 migration pathways in proteins can be found here.
Overview
Why NAMD? (in pictures)
How to Cite NAMD
Features and Capabilities
Performance Benchmarks
Publications and
Citations
Credits and Development Team
Availability
Read the License
Download NAMD Binaries
(also VMD)
Build from Source Code
- Git access now available
Run at NCSA, SDSC, NICS, or Texas
Training
NAMD Developer Workshop in Urbana (August 19-20, 2019)
PRACE School on HPC for Life Sciences (June 10-13, 2019)
"Hands-On" Workshop in Pittsburgh (May 13-17, 2019)
Charm++ Workshop in Urbana (May 1-2, 2019)
Enhanced Sampling and Free-Energy Workshop (Sept 10-14, 2018)
NAMD Developer Workshop in Urbana (June 11-12, 2018)
"Hands-On" Workshop in Pittsburgh (May 21-25, 2018)
"Hands-On" QM/MM Simulation Workshop (April 5-7, 2018)
Older "Hands-On" Workshops
Support
Mailing List Issues for Yahoo.com Addresses
Announcements
NAMD 3.0.2 Release (Aug 2025)
NAMD 3.0.1 Release (Oct 2024)
NAMD 3.0 Release (Jun 2024)
NAMD 3.0 New Features (Feb 2024)
NAMD 2.14 Bug Fixes (Apr 2022)
NAMD 2.14 Release (Aug 2020)
NAMD 2.14 New Features
One-click NAMD/VMD in the cloud
QM/MM Interface to MOPAC and ORCA
QwikMD GUI Released in VMD 1.9.3
Previous Announcements
Documentation
Related Codes, Scripts, and Examples
NAMD Wiki (Recent Changes)
Older Documentation
News
Sparing healthy microbes while using a novel antibiotic

AMBER force field use in NAMD for large scale simulation

NAMD GPU-resident benchmarks available

NAMD and VMD share in COVID-19 Gordon Bell Special Prize
NAMD reference paper published online
Coronavirus Simulations by U. Delaware Team
Coronavirus Simulations on Frontera Supercomputer
Breakthrough Flu Simulations
Oak Ridge Exascale Readiness Program
Prepping for Next-Generation Cray at NERSC
Supercomputing HIV-1 Replication
How GPUs help in the fight against staph infections
Computational Microscope Gets Subatomic Resolution
Opening New Frontiers in the Battle Against HIV/AIDS
HIV Capsid Interacting with Environment
Assembling Life's Molecular Motor
Older News Items



