Optimizing Lattice Microbes, NAMD, and VMD for Power Efficiency
Many of the continuing scientific advances achieved through computational biology are predicated on the availability of ongoing increases in computational power required for detailed simulation and analysis of cellular processes on biologically-relevant timescales. A critical challenge facing the development of future exascale supercomputer systems is the development of new computing hardware and associated scientific applications that dramatically improve upon the energy efficiency of existing solutions, while providing increased simulation, analysis, and visualization performance. Our team has developed hardware instrumentation and associated software to allow detailed power measurements to be made for computational biology applications, thereby enabling performance and efficiency of computationally biology applications to be measured and improved.
Publications
- Evaluation of emerging energy-efficient heterogeneous computing platforms for biomolecular and cellular simulation workloads. John E. Stone, Michael J. Hallock, James C. Phillips, Joseph R. Peterson, Zaida Luthey-Schulten, and Klaus Schulten. 2016 IEEE International Parallel and Distributed Processing Symposium Workshop (IPDPSW), pp. 89-100, 2016.
Power Profiling Instrumentation
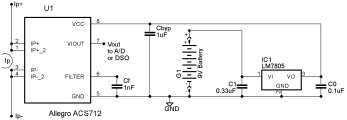
The Center has developed low-cost hardware power monitoring instrumentation, enabling developers of computational biology applications to profile power consumption with sufficient temporal resolution to measure the power characteristics of individual subroutines. This instrumentation can monitor multiple hardware sensors, which permits measurement of computer systems containing multi-core CPUs and GPUs or other accelerators. We have developed hardware power measurement sensors that can be easily interfaced to commercially-available digital storage oscilloscopes (DSOs) or to low cost network-accessible analog-to-digital converter (ADC) devices. We have developed associated software modules to communicate with both DSOs and ADC units, as well as commercially available power distribution units (PDUs) that contain built-in on-board power monitoring instrumentation of their own, enabling a variety of monitoring instrumentation to be used for measuring power consumption of computational biology applications running on mobile computing tablets, laptops, desktop computers, and small clusters.
Adaptation of GPU-Accelerated Molecular Modeling Applications to the ARM Platform
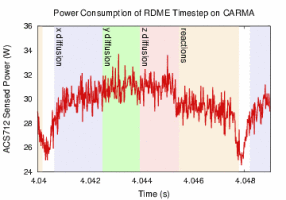
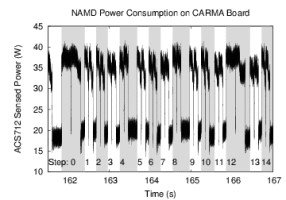
To investigate the potential benefits of emerging energy-efficient hardware architectures, the Center has ported Lattice Microbes, NAMD, and VMD to several NVIDIA Tegra development platforms based on the ARM architecture, which allows performance and power efficiency measurements to be made on emerging computing platforms in both the mobile and server spaces. All three applications also support GPU-accelerated computing, and our power monitoring instrumentation enables GPU activity to be monitored in detail.
We have begun making detailed studies of both the performance and power efficiency of key algorithms arising in molecular dynamics simulations, molecular visualization, and stochastic cellular simulations, with the goal of developing new algorithms that achieve the same or higher performance while reducing energy consumption. The improvement of the energy efficiency of computational biology applications will be critical for their success future exascale hardware platforms.
Investigators
- John Stone
- Jim Phillips
- Mike Hallock
- Joe Peterson
- Kirby Vandivort
- David Hardy
- Zan Luthey-Schulten
- Klaus Schulten