Highlights of our Work
2026 | 2025 | 2024 | 2023 | 2022 | 2021 | 2020 | 2019 | 2018 | 2017 | 2016 | 2015 | 2014 | 2013 | 2012 | 2011 | 2010 | 2009 | 2008 | 2007 | 2006 | 2005 | 2004 | 2003 | 2002 | 2001
A pendulum swinging back and forth every second due to the law of gravity is a common sight. By going down to nanometer dimensions new phenomena emerge under different physical laws. According to a recent report, a potassium ion is found to swing back and forth inside a nanoscale tube at a terahertz frequency (a trillion times a second). Unlike the pendulum, the ion's oscillation is driven by electrostatic interactions with electrons inside the nanotube wall as shown in the figure. The tube, a carbon nanotube, is composed of a cylindrical hexagonal lattice of carbon atoms; the ion induces through a so-called dielectric response charges in the nanotube wall that interact back with the ion. This dielectric response of the nanotube electrons, ordinarily, can be described only through time-consuming calculations, but based on previous work (see Jan 2005 highlight) the response can now be calculated very quickly, in effect, on-the-fly along with the ion motion. The calculations revealed that carbon nanotubes attract ions into their inside and make them oscillate at Terahertz frequency. The Terahertz oscillator may serve as a detector in future imaging devices. (See also our nanotube website).
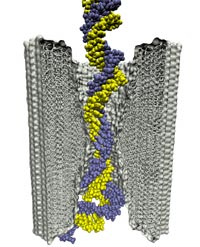
The most celebrated molecule of living cells, DNA, owes its fame to its role as a carrier of genetic information. But DNA is also impressive through other amazing properties, for example its mechanical flexibility. At first sight, it might seem a dull question to ask what is the smallest pore DNA can be squeezed through, as the obvious answer is that the diameter of that pore should be slightly larger than the diameter of a DNA helix. However, recent studies (paper1, paper2) in asking the stated question discovered that double stranded DNA can permeate, without loosing its structural integrity, pores smaller in diameter than a DNA double helix. The discovery was initiated through molecular dynamics simulations, carried out using NAMD and VMD. The simulations demonstrated that if an electrical field, driving negatively charged DNA through a nanopore, exceeds some critical value, the force exerted on DNA stretches DNA to twice its equilibrium length, reducing thereby its diameter and allowing it to squeeze through narrow pores. The simulations predicted precise values of pore radii and associated critical fields. The predictions were validated experimentally by counting the number of DNA copies that passed at different electric fields through synthetic nanopores. Further details about this study can be found here.
image size:
344.8KB
Olga Svinarski and VMD
It was 1995 when NAMD was introduced (Nelson et al.) as a parallel molecular dynamics code enabling interactive simulation by linking to the visualization code VMD. In 1999 a major improvement was accomplished in NAMD 2 (Kalé et al.), scaling to 200 processors at the time due to the efforts and software of the Parallel Programming Lab. NAMD has since matured, adding many features and scaling to thousands of processors, garnering accolades and users in the process. This progress is now collected in a NAMD review paper that presents, in a manner accessible to the novice researcher, the concepts and algorithms behind NAMD, features for steered and interactive MD and for free energy calculation, the elements of the NAMD design that enable parallel scaling, performance comparisons of a variety of platforms, and advice for productive use of NAMD on modern research projects. Case studies ranging from the typical to the elaborate demonstrate the capabilities and flexibility of NAMD. This new reference provides an excellent foundation for working through the extensive NAMD tutorials, either on your own or at a hands-on workshop.
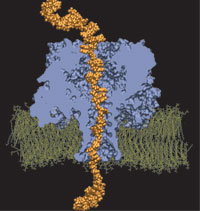
For the sequence of DNA, the genetic instructions of cells, to be read, the double helix of DNA is split open, exposing single DNA strands to DNA binding proteins. Once bound to DNA, the proteins, in carrying out their functions, will crawl along the DNA strand in one of two directions, towards DNA's 3' or 5' end. A recent study of DNA discovered a surprising property of single DNA strands that seems to explain how DNA binding proteins recognize the right direction on DNA strands. By measuring the translocation of DNA through alpha-hemolysin, a membrane protein with a narrow pore, researches discovered that directed single stranded DNA moves much faster when entering the pore 3' end first, rather than 5' end first. The underlying mechanism of this directionality was discovered through molecular dynamics simulations using NAMD and VMD. The simulations revealed that, in a narrow pore, DNA bases tilt collectively towards the 5' end, transforming a wide space directionless DNA brush into a tight space DNA ratchet. The 360,000-atom MD simulation did not only reveal how the DNA bases align and move faster in the "smooth" direction, but did also predict how the directional DNA movement can be discerned by means of direction-sensitive ionic currents through the channel blocked by translocating DNA strands. More details about this study can be found here.
image size:
102.1KB
made with VMD
In an optimistic future, cars and appliances will be powered by renewable energy produced by burning hydrogen gas, with water being the only waste product. To supply this hydrogen gas, scientists are turning their attention to an enzyme called hydrogenase that is found in certain microorganisms, which produce hydrogen gas from sunlight and water. This enzyme, however, is sensitive to oxygen gas, which irreversibly deactivates its hydrogen-producing active site. Understanding how oxygen reaches the active site will provide insight into how hydrogenase's oxygen tolerance can be increased through protein engineering, and in turn make hydrogenase an economical source of hydrogen fuel. In a recent paper (also described in this webpage), the programs NAMD and VMD are used to analyze the gas diffusion process inside hydrogenase, and how it correlates with the protein's internal fluctuations, thereby creating a map of the oxygen pathways. The calculations revealed two distinct pathways for oxygen to reach the active site. Gases participate in physiological processes of many organisms and the new computational strategy developed promises to image gas diffusion pathways for many relevant proteins. In fact, the researchers are currently inspecting hundreds of proteins for their ability to internally transport gas molecules.
image size:
313.8KB
made with VMD
Sun light is the primary source of energy for Earth's biosphere made available through photosynthetic organisms. In the cells of such organisms, so-called photosynthetic units form in which about a hundred interacting chlorophylls are assembled into a molecular network. The network (see figure) absorbs through its individual chlorophylls light and transfers the ensuing electronic excitation and with it the light energy efficiently along the network connections to a center where the energy is transformed into a membrane potential that subsequently fuels the machinery of the organisms' cells. The biological evolution that lead to such energy transfer networks had to obey physical constraints that act upon the network assembly and all stages of the light harvesting process. With the availability of atomic level structures of two networks, one in plants and one in cyanobacteria, it became possible to compare the energy transfer networks from two related species that are separated by more than a billion years of evolution. The comparison is reported in a recent paper and on our web site. It answers many questions about how living systems evolved extremely efficient solar cells for their energy needs, while still leaving a puzzle behind to be answered: despite a huge evolutionary distance the plant and bacterial photosystems maintained an amazing identity as the positions and geometry of about eighty chlorophylls remained unchanged for over a billion years. For most of these chlorophylls, repositioning would have had little effect on the efficiency of the light harvesting processes as calculations show, yet evolution chose to freeze the chlorophyll ensemble geometry-wise.
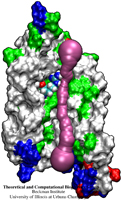
image size:
342.1KB
made with VMD
The import of nutriments over their cellular membranes is one of the main tasks of all living cells. Even though a major part of the cell's molecular machinery is devoted to this task, principles of selective membrane transport are not yet well understood, mainly due to the fact that the membrane proteins responsible are notoriously difficult to resolve in their structure, the latter a prerequisite for a full physical description of the function. Recently, cell biology got very lucky in having the structures of two closely related membrane proteins solved. Two highly homologous aquaporins from the same bacterium, Eschericha coli, have become structurally known: one that conducts only water, called AqpZ, and one that conducts water as well as the nutriment glycerol, called GlpF. The discoveries have permitted us through structure analysis with VMD and molecular modeling with NAMD to look over nature's shoulder in the evolutionary design of two similar import channels of different conductivity. As described in a recent paper and on our aquaporin site, in making a water channel also a glycerol channel, nature has turned to a very basic principle, namely adjusting the overall pore size of the channel from a very narrow channel, just wide enough for water, to one wide enough also for glycerol.
To guide cell biology research and explain observation through molecular structures and sequence data, life scientists resort increasingly to computational tools. Sequence and structure viewers (VMD) combined with molecular dynamics modeling software (NAMD) are primary methodologies that revolutionized modern biomedicine. The revolution happened so quickly, though, that traditional university training has not kept up with the pace of developments in computational biology. A series of computational biophysics workshops in Perth (Australia), Urbana, Boston, Lake Tahoe, Chicago, and San Francisco attempted to fill the gap through hands-on training. Theory sessions in the morning introduced the concepts and methods used in molecular modeling today; computer laboratories in the afternoon gave participants, students, postdocs, and faculty, opportunities to work through tutorials at their own pace on provided laptops, as well as work on their own research problems. The workshops funded by NIH, NSF, NCSA, UIUC, and UWA met the needs of novices and experts alike for instruction in a new generation of research methods. All workshop materials are available on the web.
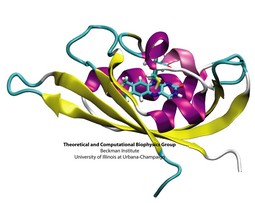
image size:
162.1KB
made with VMD
Plants and other photosynthetic organisms convert sunlight into various forms of metabolic energy. To expose themselves optimally to the sun while at the same time avoiding damaging overexposure to light, these organisms employ molecular photosensing systems that control, for example, the orientation of their leaves. Common photosensing systems include photoreceptors of the so-called phot family that are sensitive to blue light and contain Light, Oxygen, and Voltage (LOV) sensitive protein domains as photoactive elements. Light absorbed by a flavin molecule leads to bond formation with the protein (LOV domain), thereby, initiating signaling until the flavin-protein bond breaks spontaneously. A study of the photosensing events in the LOV domain of the algae C. reinhardtii (see figure), reported in a recent paper, employed computer simulations that combined quantum mechanical and classical simulation methods to study photoexcitation and subsequent processes. It emerged that formation of the flavin-protein bond is initiated by a unique light-driven transfer of a hydrogen atom between the LOV domain and the flavin molecule (see our biological photoreceptors website).
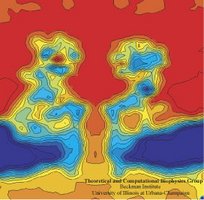
In a biological cell, membrane channels act like miniature valves regulating the flow of ions and other solutes between intracellular compartments and across the cell's boundary. Assembled in complex circuits, they generate, transmit, and amplify signals orchestrating cell function. To investigate how membrane channels work, life scientists, using an extremely fine pipette, isolate a tiny patch of a cell membrane and, in so-called patch clamp measurements, determine electric currents in response to applied electric potentials. Dramatic increase in computational power and its efficient utilization by NAMD allows one today to reproduce such studies computationally, calculating the permeability of a membrane channel to ions and water directly from its atomic structure. In what is one of the largest molecular dynamics simulation to date, described in a recent paper as well as on our web site (here), one copy of the membrane channel alpha-hemolysin, submerged in a lipid membrane and water, was subjected to an external electric field that drove ions and water through the channel. The calculations produced also an image of the electrostatic potential across the channel (see figure).
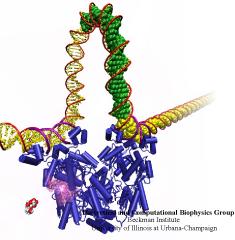
image size:
80.0KB
made with VMD
When Escherichia coli bacteria enjoy lactose and related food molecules in their environment, the cells quickly furnish proteins needed for import and metabolic digestion of the food. A set of genes, called the lac operon, is transcribed into messenger RNA that directs the synthesis of these proteins. When lactose is not available, the protein synthesis would be wasteful and, indeed, is prevented by locking the lac operon. This is achieved by a protein called lac repressor that forces the segment of the lac operon needed to initiate transcription into a loop, but that can be unlocked by a lactose molecule binding to the protein as soon as the food becomes available again. A recent study of the lac repressor combines a 314,000-atom protein simulation using NAMD with a multiscale simulation technique coupling the protein to the DNA loop. The calculations reveal how the lac repressor stretches out two "hands" grabbing the genomic DNA and then keeps a tight grip on the DNA wrestling it into a loop. The discovery is described on our website as well as in a popular article.

image size:
102.0KB
made with VMD
The ear is a sensitive and robust device, able to perceive the faint sound of flowing water and the thunderous blast of an air plane. Like a microphone, the ear transforms a complex, mechanical stimulus (sound), into an electrical signal, a voltage change in a nerve cell, that can be understood by our brain. This transformation is called "mechanotransduction" and is accomplished by a series of amazingly minute devices that each connect a soft spring to an ion channel, both located in specialized sensory cells, the hair cells of the inner ear. The springs, through their vibrations agitated by particular sound frequencies, control ion currents passing through the channels, thereby, modifying the hair cell internal electrical potential. This leads to neural signaling to the acoustic cortex of the brain. Recently reported molecular dynamics simulations using NAMD, some of the most extensive simulations accomplished to date both in size and duration, showed that the mechanical characteristics of hair cell signaling may be traced to a single protein, ankyrin, that acts as a helical spring. Imagine a soft spring that is extended several inches by the weight of a feather! Ankyrin is such a spring, but a billion times finer (see our ankyrin website).
image size:
138.8KB
made with VMD
Biological evolution left its many traces in the form of organisms as well as in "fine print" in the form of gene sequences and associated protein structures. From the "fine print" researchers can draw conclusions about the inner workings of living cells and derive opportunities to battle disease. Researchers enjoy easy access to sequence and to structure information, but so far mainly separately, i.e., either for sequence or for structure. VMD, our widely used structure viewing and analysis program, has already offered a glimpse of the viewed protein's sequence, but with its latest release has taken a key step further, assisting in viewing and aligning multiple structures and sequences with few mouse clicks. Users of VMD 1.8.3 find themselves routinely comparing their protein of interest with analogous ones getting VMD to color the protein by similarity in structure, in sequence, and showing conserved amino acids. VMD 1.8.3 surprises with numerous further features, including a new cartoon representation that follows the actual molecular structure closely and offers superb, publication quality images. VMD continues to work together with the molecular dynamics program NAMD, permitting viewing and analysis of huge trajectory files by supporting 64-bit processors.

image size:
162.2KB
made with VMD
Biological cells, the basic units of life, are organized assemblies of nanodevices. Nanobiotechnology can adapt Nature's solutions for its own purposes, using computational biology to redesign Nature's nanodevices. In the case of Nanodiscs, bioengineers thought to construct the smallest possible environment that mimics the native environment of membrane proteins. Researchers borrowed the amino acid sequence of a naturally occurring class of proteins, lipoproteins, which are involved in the transport of lipids and cholesterol. The lipoprotein found in humans, apolipoprotein A-1, was used to synthetically engineer "belts" that surround a discoidal lipid bilayer, shielding the hydrophobic lipid tail groups from water. As recently reported, molecular dynamics simulations using NAMD showed an atomic level image of the structure of such a nanodevice. The predicted discoidal shape, diameter, and thickness of Nanodiscs simulated were experimentally corroborated through so-called small angle X-ray scattering. For more details see the HDL & Nanodisc website.

image size:
199.4KB
made with VMD
An important means for generating genetic diversity to provide raw material for evolution and maintain genomic stability is sexual reproduction. At the molecular level, the genes of two individuals are mixed through a process called homologous recombination. This process is found also in many simple life forms, even bacteria. At the beginning of recombination, two DNA duplexes, e.g., from mother and father, are aligned next to one another as the result of homology search, i.e., like strands are brought together with like strands. The four single DNA strands, two in each duplex, cross reciprocally two of the strands between the duplexes. The result is a joint molecule that contains DNA crossovers, named Holliday junctions. The Holliday junction is highly polymorphic in moving along two DNA duplexes, exchanging their DNA. Researchers are now investigating the physical mechanism of Holliday junction migration. The polymorphic, dynamic character of this migration makes observations difficult and the researchers resorted to molecular dynamics simulations using NAMD. The results, reported recently, resolved the dynamics of maternal-paternal DNA exchange through Holliday junction transitions in unprecedented detail providing an atomic level view of sexual reproduction. Check a brief review on our website.
The interdisciplinary nature of modern science calls for collaborations across the campus, across the country, and even across the globe. Computer networks are ideally suited to assist researchers in this regard and this month a brand new release of the BioCoRE collaborative environment for biomedicine has taken a huge step towards getting researchers working together over small and large distances. BioCoRE sports a new, beautiful, and very responsive web interface and BioCoRE's control panel now has integrated Google® searches as well as mathematical expression solving, and private conversations with collaborators are intuitively displayed. To ensure further improvements, BioCoRE's programming interface and computer code have been redesigned to permit development of instant messaging and easier programming by third parties. All of this promises to make BioCoRE the next best thing to doing research together in the same room.
Energy for most of the earth's biosphere is gained when sun light absorbed drives electrons across a membrane through a protein called the photosynthetic reaction center (RC), leaving behind positive electron holes. The electrons join protons to become hydrogen atoms and move, bound pairwise to a quinone molecule, to another protein, the so-called bc1 complex. Here electrons and protons move together back over the membrane and become separated again, thereby establishing an electro-osmotic potential that fuels many cellular processes. However, the electrons need to return to the RC to fill the electron holes left behind. Nature employs for this purpose a kind of bioelectric extension cord in the form of a third protein, cytochrome c2, that shuttles the electrons back from the bc1 complex to the RC. A recent paper reports molecular dynamics simulations using NAMD that investigated how cytochrome c2 plugs into the RC. Landing on a broad face of the RC, interactions steer the protein such that its electron carrying heme group comes close to RC's chlorophylls with electrons missing, a chain of water molecules providing an electrical conduit. The study is yet another example of how simulations provide today complete views of the fundamental processes underlying life.

image size:
117.6KB
made with VMD
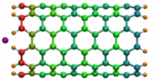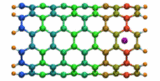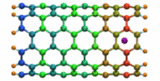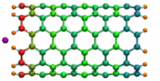
Carbon nanotubes are becoming universal tools and building blocks
in nanomedicine, being proposed as nanodevices for drug delivery,
DNA transfection, and biosensing. They can also be employed as
nanopores that conduct protons, ions, and small molecules (see June 2003 highlight), or
as reaction vessels for new types of chemical reactions. Studying
carbon nanotubes can assist in designing improved nanodevices. One
approach to studying nanotubes is furnished by molecular dynamics
simulations. However, such simulations must account for one key
property of nanotubes, their large polarizability due to the
mobility of ![]() -electrons over the
tube walls. Until recently this polarizability could only be calculated
through expensive quantum chemical calculations that could not be linked
to simulations imaging molecular processes around carbon nanotubes.
Recent studies (
1 ,
2 )
report now an empirical model that can be efficiently implemented into
molecular dynamics simulations to take into account the polarization
effect. The model reproduces results of more expensive quantum chemistry
calculations very well. A first application of the new model studied
the transport of water molecules through nanotubes. Water has a strong
dipole moment that polarizes the nanotube wall and, therefore, provides a
stringent test for the new methodology. For more informations, check out
our nanotube website.
-electrons over the
tube walls. Until recently this polarizability could only be calculated
through expensive quantum chemical calculations that could not be linked
to simulations imaging molecular processes around carbon nanotubes.
Recent studies (
1 ,
2 )
report now an empirical model that can be efficiently implemented into
molecular dynamics simulations to take into account the polarization
effect. The model reproduces results of more expensive quantum chemistry
calculations very well. A first application of the new model studied
the transport of water molecules through nanotubes. Water has a strong
dipole moment that polarizes the nanotube wall and, therefore, provides a
stringent test for the new methodology. For more informations, check out
our nanotube website.